What does body building, anti-aging cream and Bleomycin (a cancer drug) have in common? Peptides of course! Peptides are large molecules that are vital to life. If you were to take a protein and break it into smaller pieces, each piece would be called a peptide. Just like proteins, peptides are made of amino acids linked together in a chain-like structure. Whenever you ingest a protein, your body breaks it down to its individual amino acids. It then puts those amino acids back together in a different order to make whatever peptide or protein your body needs. Insulin, for instance, is a peptide that is 51 amino acids long. Your body synthesizes insulin from the amino acids it gets from the proteins you eat.
Peptides and small proteins can be synthesized in a lab as well. Peptide synthesis is a huge market in the pharmaceutical and skin care industry. They’re also used, somewhat shadily, as a steroid substitute by serious athletes and body builders. In this article, we’re going to go over the basic steps of how to join amino acids together to make a peptide. The chemistry of peptide synthesis is complex and well beyond the scope of this article. But the basic steps of making a peptide are not as difficult as you might think. Join me after the break to gain a basic understanding of how peptides are synthesized in labs across the world, and to establish a good footing should you ever wish to delve deeper and make peptides on your own.
Amino Acid Overview
There are 20 amino acids that occur in nature. They all have the same core — an amine group (NH2) attached to a carboxylic acid (COOH) via a single carbon atom, called an alpha carbon. The thing that gives each amino acid
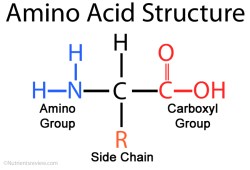
its uniqueness is the functional group, called the R group or R side chain, that is also attached to the alpha carbon. When building peptides, the amine group of one amino acid gets attached to the carboxylic acid of the other. The R functional group determines the overall shape, structure and properties of the peptide, as it will form bonds and cause the peptide to fold in on itself. By convention, the NH2 side is drawn and written on the left and the COOH side is on the right. The NH2 side is called the N-Terminus and the COOH side is called the C-Terminus.
In order to make peptides, you start with amino acids obviously. By themselves they have the consistency of a white power and are usually stored in plastic containers. Amino acids used in peptide synthesis need to have some sort of protection on the amine group to prevent unwanted reactions. This is accomplished with a molecule — the most common of which is called FMOC. This is important to note because before attaching two amino acids together, we need to remove the FMOC protecting group from the amine side of one, but leave it on the other. Leaving the FMOC on one is necessary to prevent the same amino acid from coupling with itself. For instance, if I wanted to couple A to N, how do I prevent A-A couplings from occurring? Keeping A protected with FMOC ensures we only get an A-N outcome.
The R functional groups need protection as well, but will not be covered in this basic introduction.
Solid Phase Peptide Synthesis

Solid Phase Peptide Synthesis (SPPS) was developed by a chemist named Bruce Merrifield in 1963. His technique has become the standard for making peptides in the pharmaceutical industry. It works by attaching each peptide chain to tiny polystyrene beads. This allows you to keep your peptide inside a vessel to do your chemistry. A filter in the vessel will retain the resin (with the peptide attached) while allowing solutions to be drained out.
The basic process starts with a preloaded resin, meaning the first amino acid in your sequence is already attached to your resin. Then successive FMOC removal/coupling cycles are done to make the peptide chain. The overall process goes like this:
- Remove the FMOC protecting group from the amine side of the amino acid.
- Add the next amino acid in the chain and coupling activation reagents.
- Repeat step’s 1 and 2 until the sequence is complete.
- Cleave the peptide from the resin.
Remember that the amine side of each amino acid is protected with an FMOC group. And recall that coupling amino acids together is done by attaching the NH2 side of one amino acid to the COOH side of the other. So when you remove the FMOC group and add another amino acid, the COOH side of the new amino acid gets coupled to the NH2 side of the amino acid you just deprotected.
Let’s walk through the steps of making the peptide sequence NH2-HACK-COOH. Couplings are done from C-Terminus to N-Terminus, so we start with Lysine (K) and then couple on Cystine (C,) then Alanine (A) and finally Histidine (H) We’ll want to start with the Lysine (K) preloaded onto our resin.
Step One – Deprotection
The first thing that has to be done is to remove the FMOC protecting group that’s on the first amino acid in our sequence. This is done with a deprotection solution. Generally a 20% piperadine in DMF solution is used, but piperadine is becoming increasingly more difficult to get because of its use in the making of street drugs. Alternatively, many labs now use a 5% piperazine solution in NMP. DMF is usually the main solvent used in the peptide synthesis process because it’s much cheaper than NMP, but they both can be interchanged.
We’ll need a vessel with a filter so we can submerge the resin with the deprotection solution, and then drain the solution out while leaving our resin behind. There are numerous ways to do this, but I’d recommend something like this.
A measured amount of resin is added to the vessel, then the deprotection solution. You’ll need to do the math, as the amount of resin will dictate the amount of coupling reagents needed and final product. Let it sit for a while, then drain. If you want to check your deprotection step, FMOC has a UV absorbance at 302 nanometers.
Once you’ve done the deprotection, you need to wash several times with DMF. Any piperazine left in the vessel will destroy the coupling attempt.
This process has taken us from FMOC-K-resin to NH2-K-resin.
Step Two – Coupling
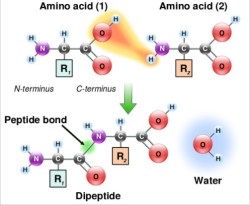
Now it’s time to go from NH2-K-resin to FMOC-CK-resin. To do this, the COOH side of C has to be coupled to the newly deprotected NH2 side of K. The COOH has to be activated before this can happen. There are several ways to do this, and every chemist has their favorite. For simplicity, I’d stick to the onium salt method. You’ll need two things — the hydrated form of HOBt and DIEA.
Solute these in DMF, as well as your amino acid, and add the proper amounts to the vessel. Again, you’ll need to do the math to know how much of each to add. Note that the ratios need to be accurate for the amount of resin, and use 3 to 5 fold excess to ensure a complete coupling.
The coupling will take a few hours. If you can use an inert gas to agitate, it will help.
Step Three – Repeat and Cleave
That’s about it really. Once you have your FMOC-CK-resin, the next step is to remove the FMOC from C and couple in Alanine (A). The process repeats over and over until your sequence is completed. Once done, you will need to remove the peptide from the resin with a process called cleavage.
The cleavage process uses a powerful acid called TFA to separate the peptide from the resin. It will also take off any R functional protecting groups. Warning – Do not handle TFA without proper protection. You’ll also need scavengers to catch any R protecting groups that come off during the cleavage process.
Crude Product
Once the peptide is cleaved from the resin, it will be in the cleavage solution. You can precipitate the peptide with very cold ether and a centrifuge. Afterwards, you’ll need access to some type of mass spectrometer to analyze your crude peptide and make sure they’re no mistakes. There likely will be, and then starts the trial and error process. Many universities will analyze it for you for a small fee.
Are you a Biohacker?
I know we have some chemists in the audience today. What do you think about a biohacker making a peptide in his or her garage? Is it possible? Realistic? Bad idea? The comments are yours, fire away.













Off Topic: I may have missed it in the past but is there an article on HaD’s resident artist (whoever did the robot at the chalkboard drawing)? Or is there more than one artist? I’d be interested in knowing their digital process and what they and the editors do for each art assignment.
http://hackaday.com/about/ [Joe Kim]
“I know we have some chemists in the audience today. What do you think about a biohacker making a peptide in his or her garage? Is it possible? Realistic? Bad idea? The comments are yours, fire away.”
Is it really hacking, or just plain chemistry?
What are the requirements to be a hack?
Do you need a soldering iron? Some spaghetti code in an esoteric language? Perhaps an Arduino? Isn’t that fake RAM ID just electrical engineering? Or any article on 3D printing, just using CAD the way it was intended?
IMO the hack comes in sourcing (more probably, synthesizing) the precursors. You may be able to find the amino acids you need for a given peptide but it’s doubtful they’ll all be in the purity you need. Lots of other reagents are controlled substances due to the drug war so you’ll have to make them or separate them from common products.
Or how about making any electronics ever? Isn’t that just what manufacturing companies do?
It’s not a hack because of the content.
“By themselves they have the consistency of a white power and are usually stored in plastic containers.”
Those racist peptides
I agree with Ostracus, how is this ‘hacking’ as opposed to just doing chemistry? The article in general is well written, however neglects to inform with common hazards and difficulties in synthesis. What is your plan for protecting yourself from the harmful volatiles that are generated from the solvents and reactions used? What is your disposal plan for the organic solvents used in SSPS? Pip, and DMF are non trivial to dispose. Also, the example you use in the intro to the article, bleomycin cannot be made with peptide synthesis, at least not economically. It is produced from a NRPS in nature, and contains non-cannonical side chains.It is basically so complicated, nature can’t even make it with ribosomes, which are the dominant form of protein synthesis. There is no obvious or solid-state route to synthesize bleomycin.
I totally agree with the caution about hazards. DMF is teratogenic, it can cause harm to the unborn child.
In terms of disposal of organic solvent, anybody who has the knowledge to do a synthesis like this probably knows how to take care of their chemicals. Personally, I distill off the solvent for reuse, and then dispose of the remaining impurities into their respective acid/base/organic waste jars.
>anybody who has the knowledge to do a synthesis like this probably knows how to take care of their chemicals
But anybody who reads this article and finds it totally new and interesting will not know how to take care of their chemicals. So they pull out their credit cards and before they know it they have a few liters of hazardous chemicals and no idea what to do with it.
Many moons ago, as a young programmer, I had to write a peptide calculator program. You can’t just jam amino acids together and expect things to work. It had to have rules about what would connect to what, and pre-generated fragments that could be inserted. I don’t remember much about it now, but I do remember it being less than trivial.
Strikes me that there HAS to be a better way to do this. Though this could be just one of those things about biology that’s pointlessly hard. My first thought is “hey, aren’t we better at stringing DNA together? Why not mix some of that with dome ribosomes?” Then it occurs to me that synthesizing DNA without massive amounts of high tech machinery is probably basically the exact same thing, but with different compounds.
The other way to make peptides is the way you, me, and that MRSA bacteria do it, and that’s with an RNA strand (copied from the DNA) run through a ribosome, The ribosome is an amazing piece of machinery, sort of a biocomputer with instructions based on six-bit word (three sets of A,C, G or T… sometimes U for Uracil in a few creatures), which map to the 20 aminos (again, a few others in different creatures but it’s remarkably consistent across species), plus other opcodes like “Stop”. RNA chains go in, proteins come out. Lather, rinse repeat.
The ribosome itself is mostly peptides, but features some pieces of RNA. it’s believed that the earliest versions were all RNA.
This is somewhat true, but the ribosome whether in vitro or in vivo is a poor way to make peptides. It obviously has great utility for recombinant protein synthesis, like cloned human insulin, but peptides do not translate well due to a multitude of reasons. Synthetic peptides have lots of advantages for use in the field. They can use non-standard amino acids, they can precisely incorporate modifications at specific positions, they can incorporate isotopes in exact positions and frequencies, and they can be quantified to exacting levels, making them useful to track and detect a duplicate version of the peptide made by nature. As for the ribosome, ATGC are the four basepairs used in DNA AUGC are used in RNA for virtually all organisms, Uracil is one of the key ways a cell uses to prevent cross-talk between RNA and DNA. The ribosome itself is an extremely complex machine and despite what you are told in school is actually mostly RNA, not protein. The ribosome is composed of about 52 proteins and 3 rRNA molecules which are much larger than the protein components. It is about 65% RNA and 35% protein.
That’s what I get for working from memory.
How do I do this in my garage though?
is there something missing from the carbon atom second from the right in the picture? (looks like it only has three things attached).
R3? Maybe it’s the last part that monitor head hasn’t written yet.
Ah, peptide synthesis! Finally something that I’m intimately connected with appears on Hackaday (I’m a peptide chemist).
To answer the final question, I doubt the chemistry’s feasible in a ‘biohacker’ context, at least not without a lot of generous donations. The actual chemistry’s not difficult – as mentioned in the comments above, sourcing or synthesising the precursors in sufficient purity would be the most difficult aspect. I’m sure someone’s seen how cheaply they can buy a kilo of cysteine or something, but bear in mind that not only does the amine need protection, so too does the sidechain if it has a reactive functional group on it (i.e. thiol in the case of cysteine, another amine in the case of lysine, etc.) The side-chain protecting group needs to survive the entire synthesis until the cleavage, so it can’t come off under the same (basic) conditions as the Fmoc group. It needs to be orthogonal. Putting two orthogonally cleaved protecting groups on a molecule can be a pain, which is why most amino acids for peptide synthesis are purchased with protection already installed.
We were recently given an old CEM Liberty peptide synthesiser for use as spare parts for our extant system. I’m sure that restoring one of those to some extent would make a great project. Getting or making the ‘ingredients’, however, would be seriously challenging for inexperienced or most hobbyist chemists.
Not to mention, many of those activating agents are pretty expensive (e.g. HATU). I rather like acid fluorides, but here in Australia the fluorinating agent of choice (cyanuric fluoride) isn’t readily available and they’re not guaranteed compatible with an automated synthesiser.
The Fmoc vs. Boc ‘debate’ is worth looking at too.
Yeah, I’m really into organic chemistry on the side, and while I’ve done some pretty lengthy syntheses, a full solid state peptide synthesis is something that I would love to attempt but is simply out of my reach as somebody with very limited access to glassware and chemicals. I really enjoyed this article though, as it’s one step closer to bringing orgo more into the hobbyist realm, which would be great for (almost) everybody.
I work at a testing lab. I don’t do the lab work… Just a glassware grunt, with a little functional brain matter. My primary interest and focus is on electronics, but I’ll admit to hearing a few familiar words that I’m not even gonna attempt to pretend I know even the slightest thing about! LOL
I’ll stick to my washing glassware and preparing incubation media broths. My brain is already overflowing with datasheets… Don’t need organic chemistry slowing down the neurons. It’s a bit over my pay grade.
Just the guy I want to talk to! So if you have a fully functioning peptide synthesizer, plus all the reagents and fmoc aminos, is that it? I can’t believe that’s all you would need. Or is making peptides that simple? I’ve been wanting to make some specific peptides, one of them is just 4 aminos long.
Is your peptide AEDG?
frank6@vcn.bc.ca
Why not just express it in piciha or e. coli? DNA synthesis is essentially eaiser you can do it with an inkjet printer, no crazy reactive side chains. Clone her into a plasmid and electroporate. Unless you wanted to make dermorphin or something that has a funky AA. At least you can have a his-tag.
My friend is doing a project on scorpion venoms and he has to synthesise the peptides with C13’s and possibly P35’s for 2D NMR. It’s pritty much taken him a year just to make it and he’s in an institution with unlimited coin…..
Recombinant for life
It’s interesting to read about solid phase peptide synthesis and how it came about. I didn’t realize that this process is as old as 1963. This is something to remember because considering how peptides are coupled and cured is very interesting.
Is there a difference in manufactured Peptides and Naturally occurring peptides? That is with regards to their effectiveness.
I don’t understand words that contain numbers and no vowels. Do chemists actually talk? Also I really do appreciate my liver.
These are another research peptide manufacturer company.