Researchers at DeepMind have proudly announced a major break-through in predicting static folded protein structures with a new program known as AlphaFold 2. Protein folding has been an ongoing problem for researchers since 1972. Christian Anfinsen speculated in his Nobel Prize acceptance speech in that year that the three-dimensional structure of a given protein should be algorithm determined by the one-dimensional DNA sequence that describes it. When you hear protein, you might think of muscles and whey powder, but the proteins mentioned here are chains of amino acids that fold into complex shapes. Cells use these proteins for almost everything. Many of the enzymes, antibodies, and hormones inside your body are folded proteins. We’ve discussed why protein folding is important as well covered recent advancements in cryo-electron microscopy used to experimentally determine the structure of folded proteins.
The shape of proteins largely controls their function, and if we can predict their shape then we get much closer to predicting how they interact. While AlphaFold 2 just predicts the static state, the sheer number of interactions that can change a protein, dynamic protein structures are still out of reach. The technical achievement of DeepMind is not to be understated. For a typical protein, there are an estimated 10^300 different configurations.
Out of the 180 million protein sequences in the Protein database, only 170,000 have had their structures identified. Technologies like the cryo-electron microscope make the process of mapping their structure easier, but it is still complex and tedious to go from sequence to structure. AlphaFold 2 and other folding algorithms are tested against this 170,000 member corpus to determine their accuracy. The previous highest-scoring algorithm of 2016 had a median global distance test (GDT) of 40 (0-100, with 100 being the best) in the most difficult category (free-modeling). In 2018, AlphaFold made waves by pushing that up to the high 50’s. AlphaFold 2 brings that GDT up to 87.
At this point in time, it is hard to determine what sort of effects this will have on the drug industry, healthcare, and society in general. Research has always been done to create the protein, identify what it does, then figure out its structure. AlphaFold 2 represents an avenue towards doing that whole process completely backward. Whether the next goal is to map all the proteins encoded in the human genome or find new, more effective drug treatments, we’re quite excited to see what becomes of this landmark breakthrough.

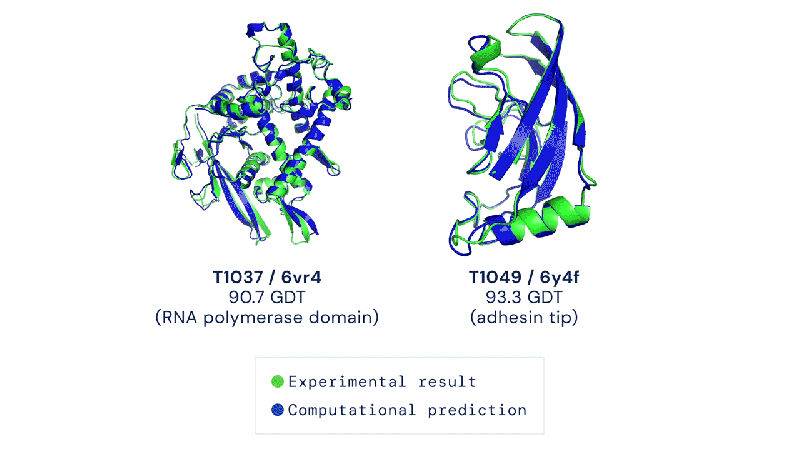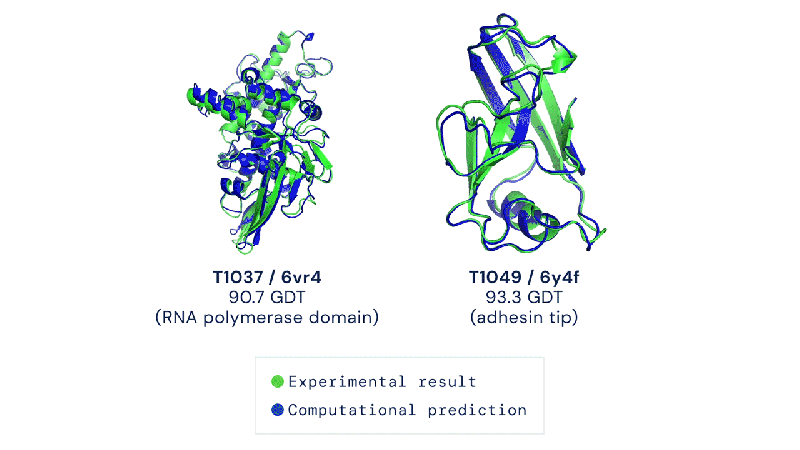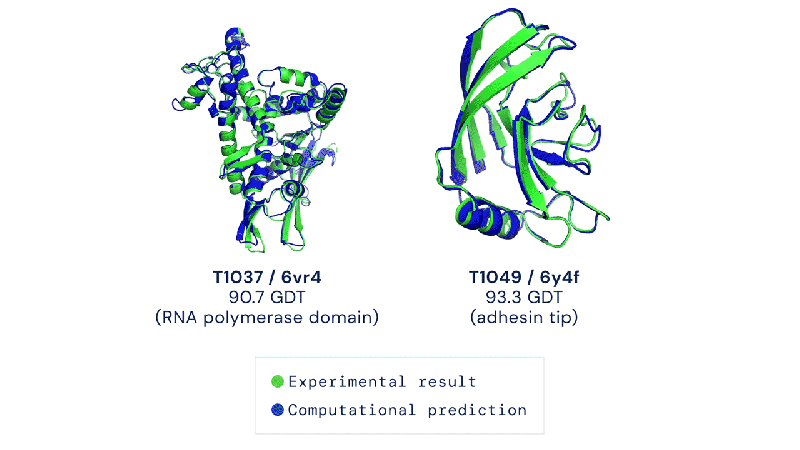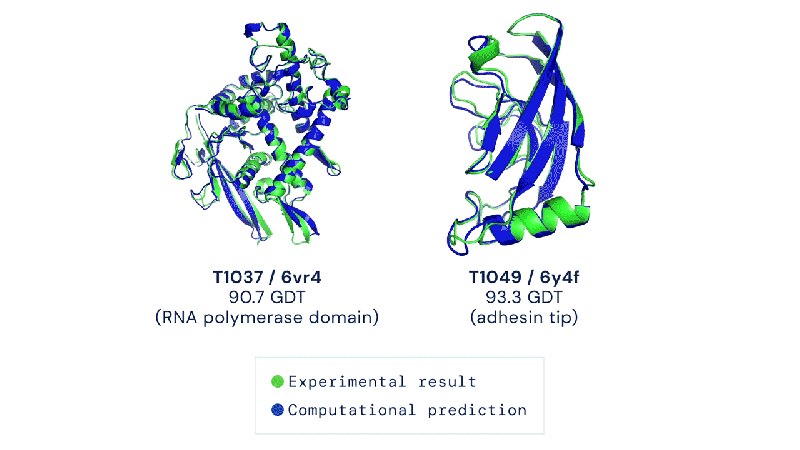













Folding at home will take on a whole new meaning.
I actually wonder how much this new folding software is complementary to projects like Folding@home, Rosetta@home etc – will it replace it?
Yes, I was thinking it will obsolete folding@home.
Nobel*
Got it, thanks.
Third line should be *Nobel* instead of Noble.
Possible that an illfolded protein might be the cause for cancers?
Proteins misfolding can cause prions, which is arguably more terrifying than cancer.
Intoxicated drivers arguably more terrifying than prions. RIP Billy Herrington.
Yep, just ask a few mad cows.
Cause? Generally no. There is evidence that prions contribute, in some cases at least, to the development of tumors, in part by inhibiting mechanisms that suppress tumor growth. See: Bom, et al, [10.1074/jbc.M112.340638], for example
All diseases are associated with autoimmunity malfunctions. Cancer is the result, proteins are the cause.
This is fantastic, and a grand achievement for mankind. However, we must not forget the historical pathway that similar breakthroughs have taken for our species. Certainly this breakthrough has power. Great power for good. Power for medicine, and many other potential benefits. It also represents a powerful attack vector against humanity. This opens the door wide open to designer proteins, that when coupled within an attack vehicle such as a virus, could absolutely devastate humanity.
Even as a passionate advocate for open source in other places, I for one hope dearly that this tool stays closed source lest it fall into the wrong hands.
Indeed.
Prions are the same folded in a different way
They lead to a cascade of other proteins flipping their conformation and give rise to dementia
This, potentially, will allow to to compare the conformation of the normal healthy protein and the pathological form
Then we can compare and choose a target site to raise antibodies to.
Previous antibody therapy failed because it attacked the normal protein too, causing massive inflammation
Like this a lot :-)
Or maybe even a direct treatment, if the prions are misfolded proteins that cause normal proteins to misfold themselves maybe this could allow us to make artificial prions that would cause misfolded proteins to refold back into normal shape? Now that would be a breakthrough!
Or a prion-like shape that has complementary topology on one side, but very different on the other, so it will cap the growing prion crystal and prevent it from continuing to grow. This would be the topology equivalent of antiviral medications like AZT and flurouracil.
(This is assuming that the issue with prions is that they crystallize into extremely long crystals, which are the nuclei of encephalopathy. There have been many suggestions for other mechanisms for encephalopathy, and even some that prion tangles are the result rather the cause of the pathology.)
I can’t help but think of the show Locke and Key while reading this article… Its like we are children, finding these magical keys that we know very little about, and we are addicted to the thrill of trying them out.
This is good news tho. Things like the whale DNA repair protein and other nature “iterated to perfection” proteins could cure lots of health problems.
Sadly, it will not cure the real human problem of Stupidity.
– Jurassic Park also comes to mind…
Kuddos! Used to love researching and advocating QSAR ( https://en.wikipedia.org/wiki/Quantitative_structure%E2%80%93activity_relationship ) and related logical ways and means for prediction. Reminded me of Deep Chem. Granted combinatorial methods are excellent too… though brute force may not be always required when there are inductive logical methods.
Amazing work but let’s not oversell it OK, there is more to the picture.
The role of molecular chaperones in protein folding
https://pubmed.ncbi.nlm.nih.gov/8529835/
The Novichok [1] family of nasties are closed source, AFAIK. Does that help?
[1] https://en.wikipedia.org/wiki/Novichok