If you are a Pythonista or a data scientist, you’ve probably used Jupyter. If you haven’t, it is an interesting way to work with Python by placing it in a Markdown document in a web browser. Part spreadsheet, part web page, part Python program, you create notebooks that can contain data, programs, graphics, and widgets. You can run it locally and attach to it via a local port with a browser or, of course, run it in the cloud if you like. But you don’t have to use Python.
You can, however, use things with Jupyter other than Python with varying degrees of success. If you are brave enough, you can use C. And if you look at this list, you’ll see you can use things ranging from Javascript, APL, Fortran, Bash, Rust, Smalltalk, and even MicroPython.
There are a few reasons I use the term “varying degrees of success.” First, not all of these kernels are official, so you never know how old some of them are until you try them. But what’s more, Jupyter works better with scripting-type languages that can run little snippets without a lot of effort. Not all of these languages fit that description. One of the worst offenders in this department is C. So naturally, I decided I wanted a C-language notebook. This is the story of how that went.
Why?
You may have many reasons for using C from a notebook. Maybe you are more adept at C than Python, and you are in a hurry. Perhaps you have some strange and wonderful library that really doesn’t want to work with Python.
You can modify those arguments to fit any of the languages you can use. However, as usual, we don’t have to figure out why to want to know how to do something. I will, however, show you a bit later that there are other options to use both Python and C in the same notebook — sort of. But first, let’s look at the actual kernel.
How?
Under Linux, at least, you have a few choices about installing Juypter. Using the repo is often unsatisfying because things aren’t there or are out of date. But if you install using the normal methods, it tends to stomp over other things you may use that have Python requirements. Ideally, this wouldn’t be the case, and sometimes everything works out, but I can tell you what has worked for me.
The first thing I do is load Anaconda, which allows you to set virtual Python environments easily. In fact, it starts with a base environment. Then I promptly turn that environment off by default. When I want to work with something like Juypter, I turn on that environment, but for normal day-to-day work, I just leave it turned off.
To do that, follow the instructions from the Anaconda website. I suggest not using sudo to install. Everything will install fine locally for your user, and there’s less chance of screwing up system software by doing it that way. As part of the installation, you’ll get a change to your .bashrc file that activates everything by default. Or you can tell it not to do that and issue a command like this to turn it on later:
. ~/my-conda-dir/bin/activate
But I just let it add a bit to my .bashrc. You need to start a new shell after you install so it will “take.” Then I immediately issue this command:
conda config --set auto_activate_base False
This won’t work if you didn’t start a new shell, by the way, so if you get an error, that’s a likely culprit. Now when you want to use the Anaconda environment for something like Jupyter, you can simply enter:
conda activate base
Obviously, if you make up different environments later, you would activate them instead. The default install has Jupyter and a lot of other things, too. However, if you want to do exotic kernels, you have some more installing to do.
For C, you want to ensure the conda environment is activated (presumably base, as I showed above). Then do the following commands:
pip install jupyter-c-kernel install_c_kernel --user
You might get a warning, but if all goes well, you are in business.
In Practice
Run jupyter-notebook, and you’ll see a start-up screen in your browser. Pick New from the right-hand side of the page and select C as the notebook type. That’s all there is to it.
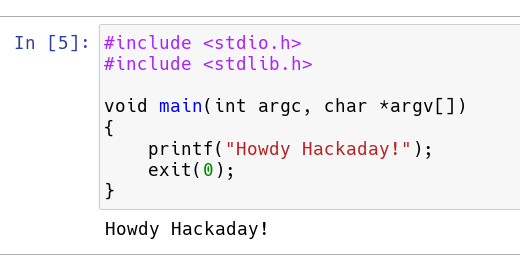
Of course, now, you need to populate a program. Whatever C code you have in a cell stands alone, so you can’t really distribute it around to different cells. It also has to be a complete compilable unit. Here’s a really simple example.
Execute in the usual way (from the menu or Control+Enter).
If you need options sent to the compiler, you can do that with a line at the top of the cell:
//%cflags:-lm
In this case, it passes the -lm option to the compiler so you can use the math library.
Other Ways
If you don’t like working in the browser, some IDEs will let you work in Jupyter notebooks. VS Code, for example, works well. Another option is to keep using the Python kernel, but compile and run your C code via the operating system. This will often even work in cloud environments that don’t support the C kernel. Naturally, too, it will work with just about anything you can do from the command line like C++, awk, or whatever.
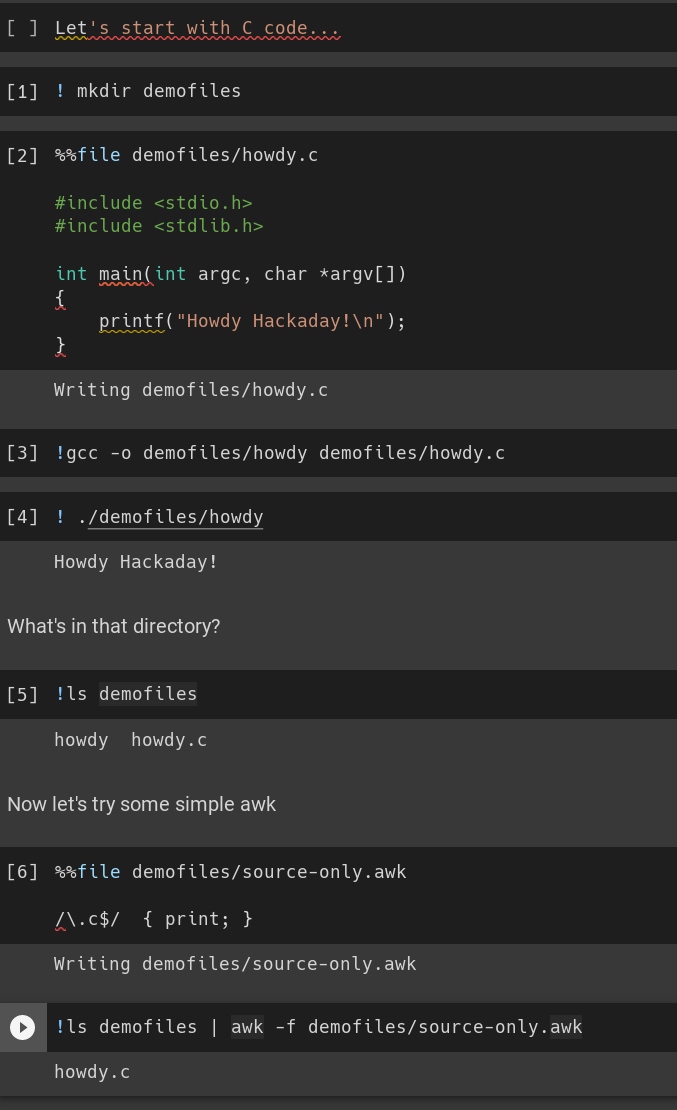
The trick is that, when using the Python kernel, you can use a ! to execute shell commands and a %% to specify that a particular cell lives in a file. So consider this notebook from Google Collab:
We make a directory, put a C file in it, compile it and run it. Later, we use awk to filter the directory listing. Nothing amazing, but you can see how you could do nearly anything with this method.
Wrap Up
Honestly, if you really want a Jupyter Notebook, C might not be your best choice for a kernel. But it is a choice, and sometimes, you really need to use some odd language from the kernel list. Maybe you have that 50-year-old Fortran code.
We looked at Jupyter, in general, a few years ago. If you prefer Fortran, we got you.














Jupyter as Arduino IDE replacement?
I am trying to use WebUSB and port some flashing tools to be able to talk to USB devices.
Arduino forked the Theia browser, but the web part is not published. Or maybe there is a way to run as a web server, being written in Electron.
I’ll continue to use Org/Babel…
I use Jupyter for micropython on ESP32 devkit and Pi Pico. Thonny crashes all the time but I have found the Jupyter version very reliable. Programming is done through local USB.
I set up the Micropython kernel some time ago on my Mac and I cant remember where I got the kernel from. When I google it, it seems that there a quite a few versions of kernels.
Al comes up with the coolest, most obscure, and sometimes weirdest Linux Foo topics. And I’m here for it. Great stuff.
I read that as A.I. lol! Maybe that’s exactly what it is ;)
“Part spreadsheet, part web page, part Python program, you create notebooks that can contain data, programs, graphics, and widgets.”
Kind of reminds me of what people tried with curl.
https://en.wikipedia.org/wiki/Curl_(programming_language)
My gut reaction was to scream “HERESY,” but to be honest, do whatever works. I might like working in the terminal with my makefiles and nerdy editor of choice, but I spent a lot of time learning that. It’s almost always better to turn diving off the deep end to get into bare metal/systems (terminal text editors, makefiles, finding that gcc fork, figuring out the flashing tool) into a nice easy wade (arduino and whatever this is). Diving in can be hard, but wading in that little bit deeper is always easy.
I use vim and the terminal as my primary programming paradigm. I would hate to try to develop in Jupyter for a significant program. But I also do a LOT of data analysis. And Jupyter absolutely rocks for that. Pulling a spreadsheet into a pandas dataframe and being able to do manipulate and plot the data with instant results in a Jupyter cell is far easier than trying to do the same thing in the terminal. Tweak a parameter or change a formula, hit control-Enter and instantly see the results in your plot. It’s damn near magic.
https://root.cern.ch/notebooks/HowTos/HowTo_ROOT-Notebooks.html
Another option for c in notebooks. Root c from cern is a nice c interpreter/dynamic compiler.
Anaconda? Multiple Python environments? Turning those environments on and off?
I’m feeling old. I am lost at how the kids are doing things these days. It’s not that I am getting stuck trying to understand how, I’m stuck before that trying to figure out the why!
Shortly before and after graduating college a couple decades or more ago I taught myself PHP. I installed LAMP on some old hardware, connected it to my always-on cable modem and forwarded the ports. My goal was to be able to reach my projects any time from anywhere. I even had an ssh terminal and a vnc client as Java applets embedded into a page so I could reach my stuff from machines I didn’t own without installing anything.
I wanted to work on my stuff without being locked in my room. Back then it was old laptops on cafe WiFi, university commons, lab and then work computers, etc… That’s mostly in my past but now with today’s tablets, smartphones and even smartphone docks that access any time, any place via the web idea is more relevant than ever. Sitting on a big rock in the middle of the woods on a nice day… sure, I can work on my projects there!
But these days… there are all these “geek” applications that are written as web apps. But it sounds like people mostly run them on localhost. They start them up manually in a terminal window then the browser and are stuck to that machine. Just the requirement of having some terminal window open that I’m not going to actually interact with in order to run a program in a different window sounds kind of cheesy. But not taking advantage of the ability to access it remotely, from any device.. It boggles my mind. Y’all have available to you exactly what I so badly wanted all those years but don’t seem to be interested at all.
And all those redundant environments, Python with Anaconda, whole systems practically with containers… why? I survived the early days of RPM Hell, ever searching rpmfind.net for that one missing library. I made it through the times when apt would uninstall half your system during an update if you didn’t read all 10,000 lines of “fine print” before hitting the ‘y’ key. It seems like just as we started getting dependency management and conflict resolution mastered, no longer a big problem a new generation came along and said “f that, I’m just going to install a whole separate environment for every program”.
I don’t get it. Help me try to get it.
We’re from the same era. The world became complicated. “fprintf()”
In 2000 a coworker showed me something early from his friend at different startup. It was “running two operating systems at once”. I said “so dual boot”. He said “no, they’re running /at the same time/”. I thought to myself “who needs that?” That startup was VMWare. Whoops!
A few years later I was working in malware detection and we routinely used it to put a condom on a computer so we could run malware safely to study it. Later I got tired of having to re-install an ocean of applications whenever my motherboard failed. I wanted to have all that stuff in a ‘bundle’, and abstracted away from the hardware which then introduced various driver issued. Computer croaked? Who cares? It was just a host machine. My real machine is a mondo file which I back up and can immediately deploy on replacement hardware that might be appreciably different that the prior. Later I would use this technique for development environments. When I onboarded folks to work on some aspect of the system, they simply had to run a copy of the pre-set-up dev VM. No shuffling through klunky documentation and spending days setting up the dev box. I still use this today for my personal stuff and also contract jobs. I keep all my clients’ work in separate VMs.
But times moves on.
Now it’s more about containers, which are like ‘lightweight VMs’, though I think that ‘souped-up chroot jails’ is more accurate. Containers do not have a full OS installed and are not virtualized hardware so are considerably smaller and performant. OTOH, they suck at a native GUI, like Windows stuff. It’s pretty much CLI or web interface for GUI in practice. But containers are awesome for modularization of subsystems. This is important in the modern era of ‘cloud’ stuff. (I did a project here where I migrated my failed server to containers for the various services I usually ran directly on the host OS. It wasn’t a particularly difficult thing, but the exercise was to kick the tires on the tech as much as the motivating disaster recovery action.)
As for Python virtual environments. Python is old. Python has a rich community where you can download wonderful modules that provide functions. However there are incompatibilities between them, and if you do much Python development for whatever reason, you’re going to get bit. The environments is weaker form of isolation involving changing environment variables and storing relevant stuff in different directories. I had occasion to use Python for some ML work at a (failed) startup. I am not a Python programmer. I am a user. And if the ML community likes Python, then that’s what I’m using for the job. But I dislike one job/project requiring something (2.7? bletch. some goofy module? barf.) that pollutes the global space for other projects. Again, I am not a pythonian, but I would suggest that one always work in a distinct environment. And if you never need to switch, then ‘who cares?’ it took only a moment to set it up to begin with.
As for Anaconda, that is slightly different. That is a package that includes Python but also an ocean of other modules meaningful for ML work. Believe me, it is a huge boon to have all that stuff there in one lump than manually install the myriad components individually just to get in position to /start/ to do your work. I had occasion several years back to do something that involved tensorflow and some opencv. I would have stalled for maybe a month or so just to figure out what things I needed and how to install them and what the dependencies were. Instead, I could install Anaconda, and a couple other things I wanted to try, and get up and boogie on the real work which was to develop some CV algorithms for drone mission planning at the edge. I am not a data scientist. I am not a CV specialist. Frankly, I’m not really anything in particular. I’m just an engineer that solves problems. We use the tools available.
The stuff we’re trying to do today is complicated, and we need tools to manage that complication. But even in the old days we had things like “fprintf()” and “fopen()”. Those managed the complications of formatting things and squirting them out to disparate hardware in a uniform way. Being freed from hardware-specific concerns, those abstractions freed us to solving problems at-hand. Super important for a commercial enterprise, and delightful for a casual user.
It’s all just a little bit of history repeating. E.g.
https://www.youtube.com/watch?v=yzLT6_TQmq8
“Super important for a commercial enterprise, and delightful for a casual user.”
Maybe my problem is that I fall in between those. This way of doing things is not delightful for me at all!
I like my home directory on a backed up NAS and my executables (binary or interpreted) on the local hard drive. I can re-install all that software after a catastrophy. And I don’t mind installing it separately on each device. Pretty much everything I use is free and the Internet is fast.
Things that sit in the home directory rather than install end up on the NAS where they run slower and waste backup space. Yuck! Several copies of the same damn version of Python and all it’s standard libraries sitting on my NAS.. makes me angry.
I’ve kind of hacked around this by creating a second, local home directory on my Desktop, moving flatpaks and other non-installable crap there and adding symlinks. But that’s ugly and really sucks to have to do!
Then there are the fact that getting a containerized app to run when the container system doesn’t well support your distro… I haven’t had dependency or conflict problems installing things direct in ages but trying to shoehorn in a container system that my distro doesn’t normally come with… Way harder!
And when a containerized app finally does work.. is it sandboxed? If I want to open a file with the app that didn’t come with it will I see my file tree or some useless chroot jail? Likewise if I want to save the data files an application generates to back them up can I easily get to them or are they 10,000 layers deep inside some obscure format of archive file?
Sorry, not sorry. This new stuff sucks. It makes everything worse. If developers released everything installable both with or without the container system that would be fine. Everyone can have it their way. But an increasing number I am noticing do not.
Wait.. Python 2.7?
< Python 3 = move on for me…
Of course, no one is paying me to use any Python apps. I'm in a Mickeysoft shop by day, hobby stuff is all Linux & Android.
For those interested in simulating systems with block diagrams in a GUI, we are working on a software called muphyn at work. It works standalone but it also runs from an ipython shell or even a jupyter notebook, allowing it to work a lot like the MATLAB/simulink duo.
It’s still alpha and crashes some times but I know for a fact that my colleage would love some bug reports ;) https://gitlab.com/CeREF/muphyn
I don’t get it… is it like a single document that contains code, documentation, photos, and executable-output? I’m visualizing something a bit like Maple, the math program?
Yes. That is a good analogy. But instead of doing math, you’re doing Python (and here, something else).
Or Mathematica?
Yeah that is so FU: C ON useful if you are into working in a literate manner.
If you’re using Anaconda just for virtual environments, Anaconda is overkill, by a lot, and miniconda will work just fine. Or use just venv, which comes with Python.
Using Jupyter in Visual Studio Code is pretty easy – just install it from the extension manager.
Anaconda _is_ overkill.
But so is VSC. Just use vim. :)
Ugh, more Python… never will understand why people want to use it.
this is about C not python. says it on the title :D