Using technologies like electron microscopy (EM) it is possible to capture molecular mechanisms in great detail, but not when these mechanisms are currently moving. The field of cryomicroscopy circumvents this limitation by freezing said mechanism in place using cryogenic fluids. Although initially X-ray crystallography was commonly used, the much more versatile EM is now the standard approach in the form of cryo-EM, with recent advances giving us unprecedented looks at the mechanisms that quite literally make our bodies move.
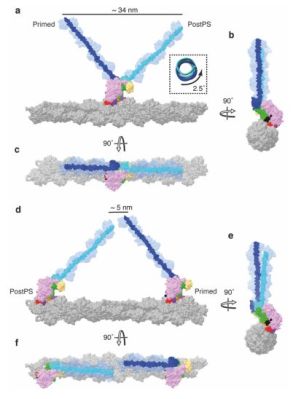
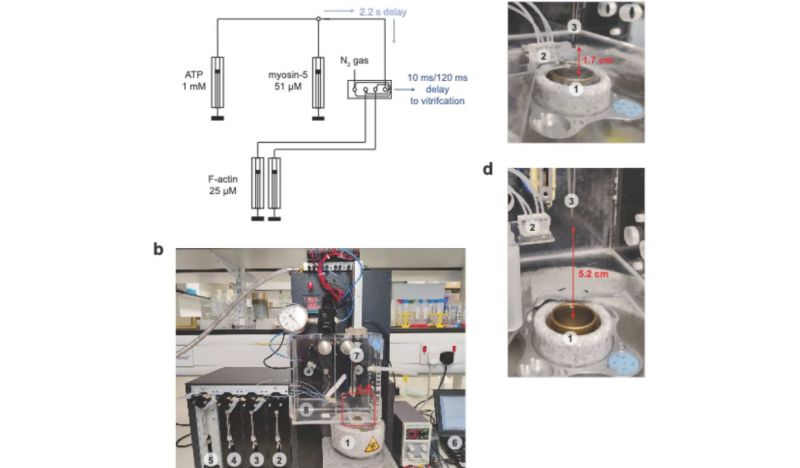
The past years has seen many refinements in cryo-EM, with previously quite manual approaches shifting to microfluidics to increase the time resolution at which a molecular process could be frozen, enabling researchers to for example see the myosin motor proteins go through their motions one step at a time. Research articles on this were published previously, such as by [Ahmet Mentes] and colleagues in 2018 on myosin force sensing to adjust to dynamic loads. More recently, [David P. Klebl] and colleagues published a research article this year on the myosin-5 powerstroke through ATP hydrolysis, using a modified (slower) version of myosin-5. Even so, the freezing has to be done with millisecond accuracy to capture the myosin in the act of priming (pre-powerstroke).
The most amazing thing about cryo-EM is that it allows us to examine processes that used to be the subject of theory and speculation as we had no means to observe the motion and components involved directly. The more we can increase the time resolution on cryo-EM, the more details we can glimpse, whether it’s the functioning of myosins in muscle tissue or inside cells, the folding of proteins, or determining the proteins involved in a range of diseases, such as the role of TDP-43 in amytrophic lateral sclerosis (ALS) in a 2021 study by [Diana Arseni] and colleagues.
As our methods of freezing these biomolecular moments in time improve, so too will our ability to validate theory with observations. Some of these methods combine cryogenic freezing with laser pulses to alternately freeze and resume processes, allowing processes to be recorded in minute detail in sub-millisecond resolution. One big issue that remains yet is that although some of these researchers have even open sourced their cryo-EM methods, commercial vendors have not yet picked up this technology, limiting its reach as researchers have to cobble something together themselves.
Hopefully before long (time-resolved) cryo-EM will be as common as EM is today, to the point where even a hobby laboratory may have one lounging around.













How tailored does the solution need to be towards the problem? That could limit commercial designs.
I would have never expected to give a post like this here… but here we are!. I am one of the main developers at cryoWrite (and a regular visitor of Hackaday). I invite you to contact us. Let us know what your specific requirements are. If we have not covered them yet, we may consider including them in a future module. Congrats for the post!
It should be noted that Cryo-EM only recently became a feasible method in the 2010s because of drastic advancements of electron cameras. This is of course a reminder that the better your science, the better your science.
For those who are interested in Cryo-EM in the UK: https://www.diamond.ac.uk/Instruments/Biological-Cryo-Imaging/eBIC.html
We have five Titan Krios microscopes, a Talos Arctica, two Glacios’s, a Scios, an Aquilos cryo-FIB/SEM, and a Leica CryoCLEM.
I am the Facility’s Manager – happy to answer technical related questions – I’m no scientist :)