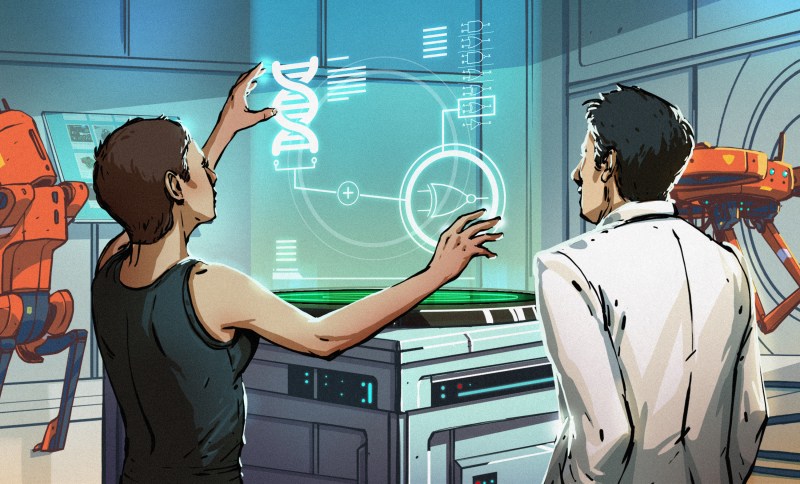
Did you know you can build fundamental circuits using biological methods? These aren’t your average circuits, but they work just like common electrical components. We talk alot about normal silicon and copper circuits ‘roud here, but it’s time to get our hands wet and see what we can do with the power of life!
In 1703, Gottfried Wilhelm Leibniz published his Explication de l’Arithmétique Binaire (translated). Inspired by the I Ching, an ancient Chinese classic, Leibniz established that the principles of arithmetic and logic could be combined and represented by just 1s and 0s. Two hundred years later in 1907, Lee De Forest’s “Audion” is used as an AND gate. Forty years later in 1947, Brattain and H. R. Moore demonstrate their “PNP point-contact germanium transistor” in Bell Labs (often given as the birth date of the transistor). Six years later in 1953, the world’s first transistor computer was created by the University of Manchester. Today, 13,086,801,423,016,741,282,5001 transistors have built a world of progressing connectivity, automation and analysis.
While we will never know how Fu Hsi, Leibniz, Forest or Moore felt as they lay the foundation of the digital world we know today, we’re not completely out of luck: we’re in the midst’s of our own growing revolution, but this one’s centered around biotechnology. In 1961, Jacob and Monod discovered the lac system: a biological analog to the PNP transistor presented in Bell Labs fourteen years earlier. In 2000, Gardner, Cantor, and Collins created a genetic toggle switch controlled by heat and a synthetic fluid bio-analog2. Today, AND, OR, NOR, NAND, and XOR gates (among others) have been successfully demonstrated in academic labs around the world.
But wait a moment. Revolution you say? Electrical transistors went from invention to computers in 6 years, and biological transistors went from invention to toggle button in 40? I’m going to get to the challenges facing biological circuits in time, but suffice it to say that working with living things that want to be fed and (seem to) like to die comes with its own set of challenges that aren’t relevant when working with inanimate and uncaring transistors. But, in the spirit of hacking, let’s dive right in.
My First BioCircuit™
Following the Composition Model (even though the Network Layer Model might be preferred), I’m going to go through the construction of a simple oscillator through electronic and biological methods. The goal is by the end you know a bit more about biological circuits, what they’re made of, what they can do and why they’re hard to make.
DNA/Physical Makeup
In electronics, we make our parts (for the most part) out of copper, silicon, and epoxy. In biology, the base parts are strands of DNA3. For this reason, if you need a quick refresher on DNA, take it. But, all you must know for this article is that DNA encodes genetic information.
Part List: Protein Expression Mediated Logic
For our oscillator, we’re going to need three inverting gates. In the electronics world, that’s pretty easy (like $0.06 easy). You can make electronic logic gates out of (almost) anything and there are countless resources detailing every… intricate… detail that any sane person would want to know.
The biological world has its fair share of logic gates, some of which we’ve already covered. We’re going to focus on the most common type: protein expression mediated logic. Through this method, protein concentrations are treated as signal and are used just like electrical signals are. Below is a genetic NOT gate, signal A (a specific protein for the Promoter or “input”) inhibits the output of signal B (the protein that the “Gene of Interest” codes for). This works just like a traditional NOT gate, if there is not signal A, then produce signal B. Well, it works almost the same way as a traditional NOT gate. This is where we find one of the great challenges that synthetic biology faces: orthogonality.
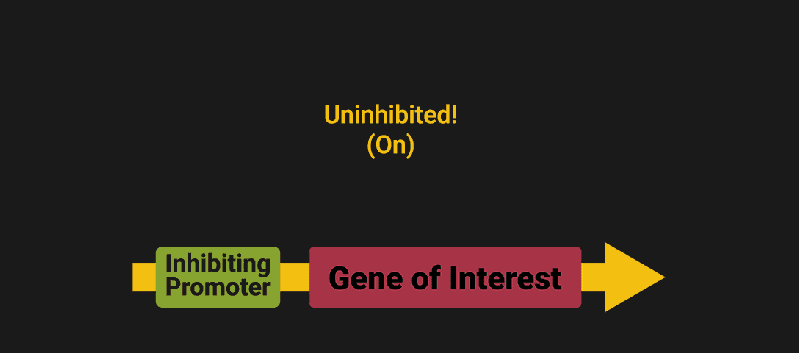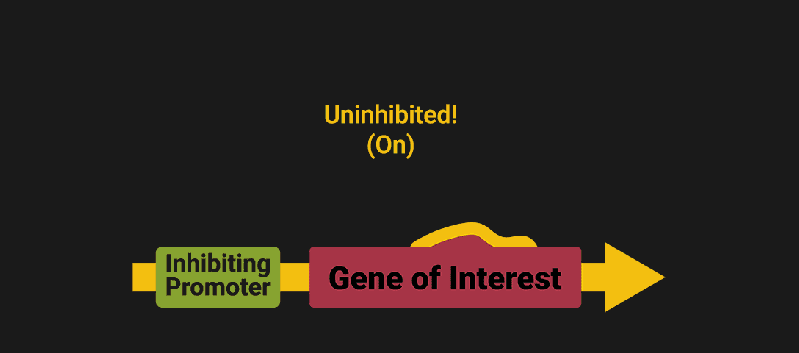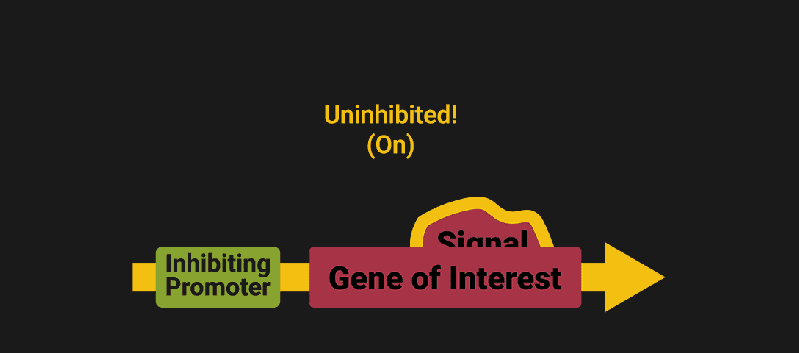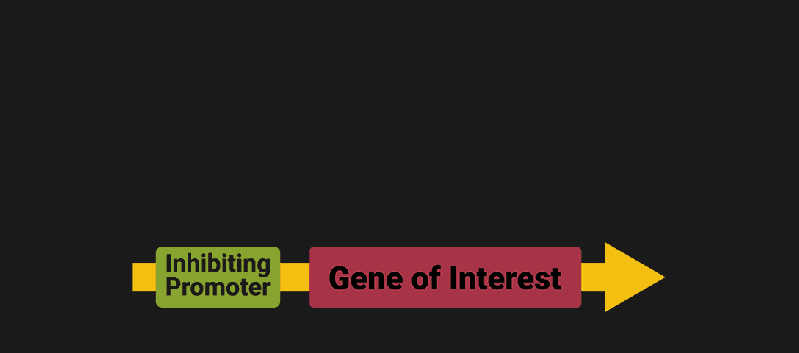
In electrical logic, we don’t usually need to worry about orthogonality, as we can connect the output of a NOT gate to what we want (and only what we want) with wires and/or traces. In biology, we don’t usually have this luxury as we’re working in prokaryotes (cells without nuclei) which can be effectively thought of as bags of chemicals4 So, instead of having direct connections from one gate to another, we only have one signal “net” as part of our network.
Computers have this figured out with identifiers like MAC addresses. For example, if Bob wants to send a message to Sue, he might broadcast something like “FOR: SUE, MESSAGE: HI”. This works out great for computers: they craft their own receivers and transmitters. In code and hardware, it can be trivial (that’s part of the reason why we have so many electronics communication standards ) but, in biology protein engineering is hard, and it takes time, ingenuity and labor.
Imagine that you could only use one type of transistor in each project. As an example, consider that if you used two 2N3904s, you turn both on or off, but never one off and one on. Now, pretend that for each transistor, you had to corner a computer in the wild, disassemble it and hand remove all of its components in the hopes of finding a new type of transistor. Surely, that would make creating a computer with 30 billion transistors, in one word, difficult.
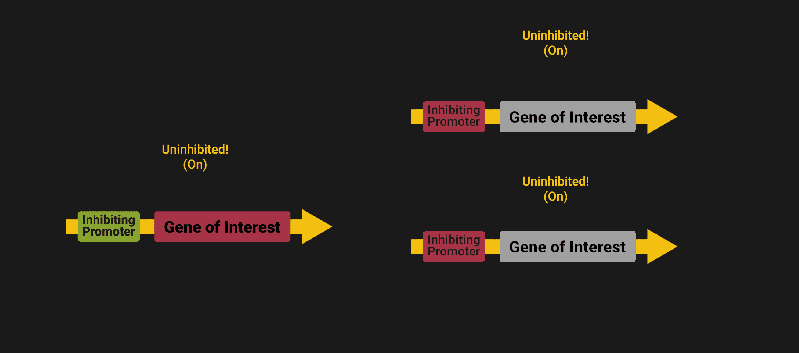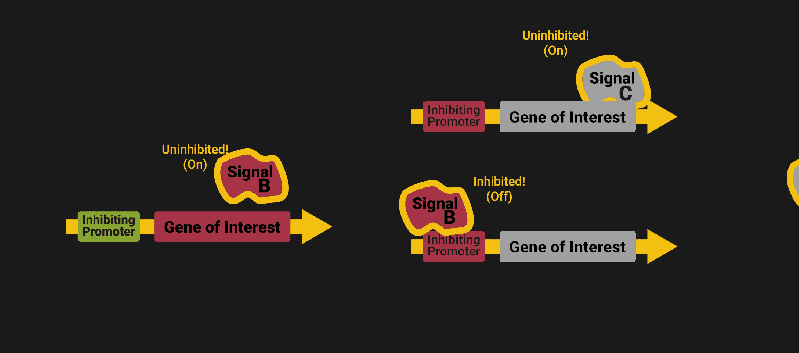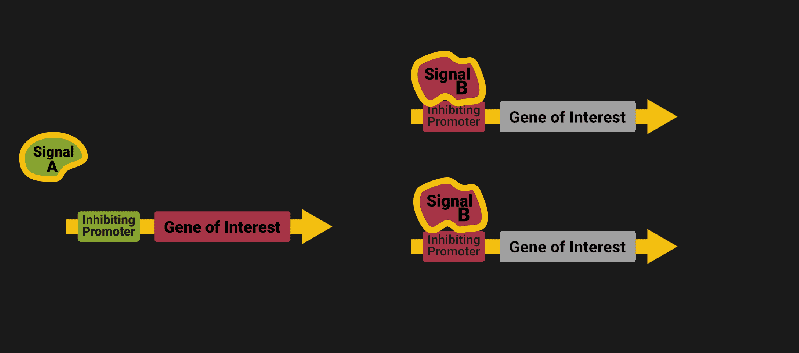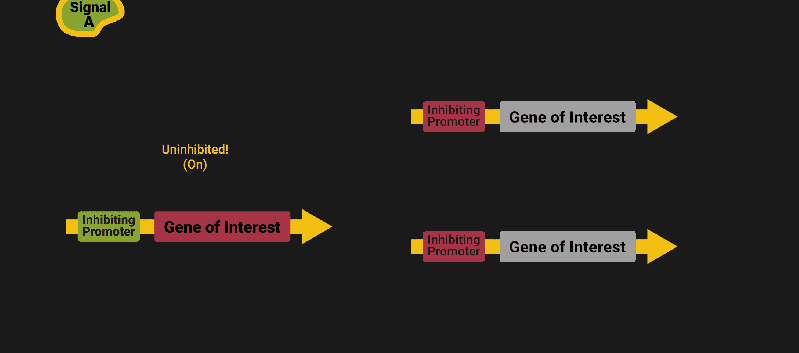
As such, practically all of the promoters5 (input sites, coded in DNA) that are used today were originally discovered in nature and have been adapted for synthetic use. And, we’ve been pretty successful at this, with enough usable promoters that we need a catalog to sort them all out6.
Device Built with NOT Gates
Now, ever forward. Let’s try to build a device with our NOT gates. And, as we’re really just here to learn concepts, I’m going to show you how to create a ring-oscillator. If you’re a bit rusty on your terminology, or you’ve been too busy to bother with a 100 level circuits class, no worries. A ring-oscillator is really simple, which is why it was one of the first bio-oscillators made. Ring-oscillators are made by placing an odd number of inverters (usually greater than 1) in a cycle (ring). See below for a very cool gif™ that hopefully should clear up any doubts about how it works. But, if things aren’t crystal clear, Enrique from MITx has your back.
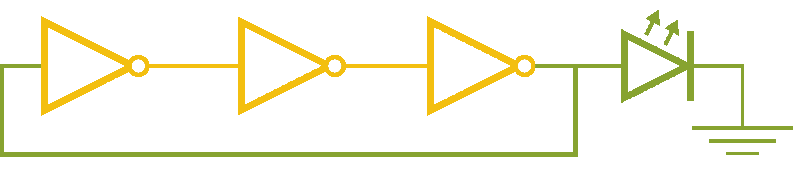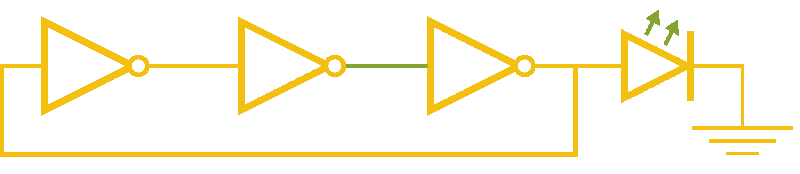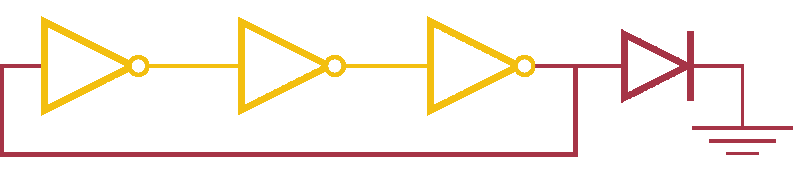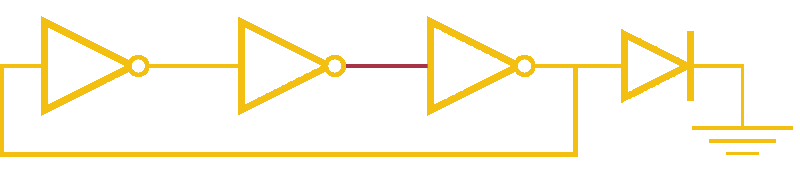
Now for the bio part. We already have our NOT gates, so surely we can string three identical NOT gates together and end up with an oscillator, right? Well, as we covered earlier, we’ll need to use three different gates, so things look like three NOT gates in parallel, rather than in series. But, even with three different NOT gates, we might have another problem: death.
After all, all our logic is taking place in a cell using up resources and producing a “useless” protein. And, by the very nature of cells, there’s a limit to the number of resources a cell has. We can try to improve our cell’s effective throughput, but at some point, we need to realize that the cell, as greedy as it is, must take care of the basic requirements of life. However, fear not. Our three gate oscillator should be sustainable7. But, if we were to try seven or even nine gate oscillators we might need to start worrying.
Anyway, enough dilly dallying. We’ve put our DNA sequences together, introduced them into a cell and now we wait for them to grow8. We’ve used GFP (a Green Fluorescent Protein) as one of our “signals” so we can visualize our signal (just like one would use an LED to view a digital signal).
And… nothing. At first. After about 160 minutes our cells start to glow. Just like electronic ring-oscillators, our frequency depends on the delay time of each NOT gate. In electrical contexts, this gives us frequencies commonly in/above the kHz range. In biology, we’re working at a blazing fast 1×10[-4] Hz with a period of ~160 minutes. Again, another stark difference between biological and electrical circuits.
In Practice / Why You Should Care
Okay, I know how it seems. Biological Logic is underdeveloped, slow and constrained by life. But, there is a harmony! What biology lacks, it makes up for in immeasurable ways. After all, we’re made of cells, not silicon.
From regulating transgene expression for regenerative medicine to a tunable dual-promoter integrator for targeting of cancer cells, cellular logic has the opportunity to change the way we think about how medicine should work, what we think life is and whether or not plant’s should glow. You should care because it’s cool it’s new and it has the potential to help lots of people. We’re hackers. We’ve always been at the forefront of technology, and just because this one’s squishy things don’t need to change. So, let’s get hacking!
I really enjoy writing about synthetic biology, as you might have guessed, so let me know if there are any topics that y’all would like to hear about. Would a few “Getting Started” articles on, well, getting started in synthetic biology with actionable procedures be a good place to start?
Notes
[1] Calculated using the figures from the Forbes Article “How Many Transistors Have Ever Shipped?”. While I have little confidence in this article, I am willing to believe that the figure presented is accurate to at least a few orders of magnitude. Regardless, the exact number is less important than the overall impression that many transistors surround us today.
[2] Specifically, the E. Coli was controlled by Isopropyl β-D-1-thiogalactopyranoside (IPTG) which is a molecular mimic of allolactose, a lactose metabolite. IPTG is used because, unlike allolactose, IPTG is not hydrolysable by β-galactosidase, so its concentration remains constant as it’s not broken down as readily.
[3] Okay, yes: DNA is made of nucleotides but copper is made of atoms and we have to stop somewhere. In our context today, it makes sense to stop at DNA, but in other contexts, such as DNA synthesis, DNA clearly isn’t the base unit.
[4] To say that cells are just “bags of chemicals” is a gross oversimplification analogous to calling our bodies just “skin with stuff inside”. Technically true, but not very descriptive. What I’m trying to get across here is that cells are (1) bound by a membrane, that they are (2) one system of transmitters and receivers and that (3) they have a limited rate at which they can absorb/deport things. This metaphor makes it easy to visualize these attributes, so I have chosen it in the spirit of Upaya (where something may not be ultimately “true” in the highest sense, but it may still be an expedient practice). For a more accurate representation of cells, check out this award winning book: Cells, Gels and the Engines of Life.
[5] Throughout this article, I use Promoter as a broad phrase, to refer to Repressors and Promoters for two reasons: simplicity and to avoid the constant use of promoters/repressors throughout the article, promoter is already a long enough word.
[6] Granted most, if not all of these promoters are only characterized for E. Coli. But, with E. Coli being the workhorse of synthetic biology, this usually isn’t an issue.
[7] By sustainable, I mean that the cell shouldn’t die from expressing the proteins that we need for our circuit. I, however, did not mean that the output would be sustainable over extended periods of time. As time progresses, our system will break down we’ll eventually get weak expression of all 3 proteins in our system. Oh well, that’s life in the city.
[8] I dislike the fact that I fast forwarded past this process. The process of getting DNA ready (not to mention preparing/designing the DNA) and coercing the cells to express your DNA is a whole ‘nother bucket of worms. Could easily be another article (or set of articles). (Wink wink)












I thought that “bio-circuit” was only a lab thing, difficult to do at home, so yes, a big yes for getting started articles!!
“[4] To say that cells are just “bags of chemicals” is a gross oversimplification analogous to calling our bodies just “skin with stuff inside”.”
“Ugly bags of mostly water”
“Home Soil”
As a biologist, I agree that describing cells as “bags of chemicals” is totally appropriate.
Fascinating! Thank you for this great article.
Yeah dude enough with the eating and sleeping and whatnot… get on with the next articles!
Totally agreed!
Synthetic biology is missing hands-on “Getting Started” articles
Nature has had a temperature driven switch for quite a while.
I’ve often wondered (half seriously) if this had any sort of ancient evolutionary link to the thermostat battles between (human) men and women! ;)
http://articles.chicagotribune.com/1986-06-29/news/8602160365_1_eggs-alligator-hatched
https://en.wikipedia.org/wiki/Temperature-dependent_sex_determination
Yeah, a getting started guide/article would be neat. However to get something actually useful it schould have (or link to) a list with suppliers to get all stuff needed (bonus points for affordability/price and ease of access). Especially considering hackadays global reader base. So lists for different continents would be needed.
THAT would actually be useful!
If it’s to much work for a single article writer, thing about converting it into a community project (e.g. hackaday.io project) to leverage the community knowledge.
In a physical world, knowledge is only a half of the required things needed to create a physical object. The rest is matter. Quit specific matter in this case.
Well, I think you get the gist.
Really interesting article about building fundamental circuits using biological methods.
I see similarities with a simulation program that I created. It is a 3D, real-time simulation of hypothetical brain cells.
The simulation runs within your Chrome browser, (it needs a powerful GPU in your pc).
You can create cells, and create connections between those cells. Cells can also execute instructions that change connections between cells and change chemicals (neurotransmitters) in the cells. There are only 3 types of instructions.
You can program the cells to perform any function, just like a computer. Examples show the generation of chess moves and a calculator that adds numbers. The project is called NeuronZoo.
See http://www.neuronzoo.com or hackaday.io/project/19287-neuronzoo
OK, one thing this article left out for me, how do you turn a signal off? I get that the presence of a given protein stops the PRODUCTION of another protein, all that has been produced is still in solution, right? Do the proteins break down on there own? Is there control of enzyme production to break them down?
perhaps my logic is flawed, but if presence of a protein is analogous to a logical “on”, than production of that protein is enough to produce that state, but what causes the protein to cease to be present, just because it has ceased to be produced?
You’ve got a good point: the proteins that are expressed need to “go away” for the signal to go low. Proteins can be very long lived (~a month or more) or have fast and furious life (~11 minutes). There are many factors that can influence their breakdown, some of which are active (eg. the cell breaks the protein down on purpose) and some of which are passive (eg. oxidative damage). Most of the factors depend on the stability of the proteins produced (influenced by lack of stabilizing ligands among other things) so one can tune the fall rate to suit the intended application. More reading if you want it: https://www.ncbi.nlm.nih.gov/books/NBK22600/#_A3198_ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3008417/ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3625778/
you are asking the right questions
The article indicates the following reaction in the first example:
thisDNAsite [+ omitted transcription reagents] -> thisDNAsite + SignalB
SignalA + thisDNAsite -> thisDNAsiteInhibitedBySignalA
in practice most chemical reactions occur in both directions (but often at vastly different reaction rates)
so there is also:
thisDNAsiteInhibitedBySignalA -> SignalA + thisDNAsite
so SignalA occasionally attaches and detaches from the inhibiting promotor.
As Michael replied, there are reactions that would also breakdown Signal A
SignalA + [omitted reagents] -> BreakdownProducts
‘if presence of a protein is analogous to a logical “on” ‘, correct: (apart from the specific DNA sites) the reagents typically take on reagent counts >> 1 per vacuole or cell. So the boolean logical gene regulatory networks are heuristic descriptions of the functionality. The analogy with analog electronics is more accurate than the analogy with boolean circuits. Consider the following reactions:
1: DNAsite + […] -> DNAsite + SignalOut
2: SignalA + DNAsite thisInhibitedDNAsite (notice double arrow)
3: SignalA + SignalB -> SignalC
and following (medium slow) decay reactions:
4: SignalC -> breakdown products
5: SignalOut-> breakdown products
if reaction rate constant of [3] is much slower than those of [1] and [2] then SignalOut = NOT(SignalA) and a boolean description is faithful.
if reaction rate constant of [3] is much faster than those of [1] and [2], then SignalOut = NOT(CMP(+SignalA-SignalB)=CMP(+SignalB-SignalA), where CMP is an analog comparator: i.e. for non-boolean concentration values of SignalB and SignalA, any common concentration is reacted away to SignalC, and there will be either only SignalA or SignalB left. This shows how reaction kinetics (rate constants) can totally modify the functionality of identical reaction schematics (where the reaction constants are unknown) i.e. omitting (or not knowing, not having measured, having no access to a database of ) reaction rates leaves us in the dark if the SignalB is like a boolean NOT gate, or if it is like an analog comparator CMP.
The theory and software for modeling such reactions has been independently written and implemented hundreds of times by different groups (look up for example Gillespie algorithm), but they are pretty useless without an open database of actual reaction kinetics (which is valuable and expensive to determine information).
Just like one shouldn’t measure component values in circuit, these reactions have to be reproduced in vitro so their reaction rate constants can be determined. Then we can faithfully simulate a large fraction of cellular behaviour (with the theory and software that is already around for decades).
To better cement the analogy between analog electronics and reactions perhaps an exhaustive table of 2 columns where each row is a concept would be useful for interdisciplinary communication between analog electronics and biochemists..
for example
biochemistry – electronics
reagent – conductor/net
reaction – component
…
@Michael: fantastic article, congratulations
thank you both for the added information.
I guess an oversimplification would be a digital circuit with crazy big capacitors between gates(so that there is a huge lag between an output going low, and it’s related input going low, but it eventually gets there).
and an oversimplification of biochemistry would be regular chemistry with reaction sites tightly wrapped in giant, relatively nonreactive molecules, so that shape plays an important roll of reactions.
PLEASE stop posting full articles to the main page with no break! It’s making me stop reading HaD :(
You can build ANYTHING given enough NAND gates…
Do octopuses really have nine separate brains?
I generally think of them having a central ring brain, with partially autonomous ganglia within each arm.