Our digital world is so much more interactive than the paper one it has been replacing. That becomes very obvious in the features of Jupyter Notebooks. The point is to make your data beautiful, organized, interactive, and shareable. And you can do all of this with just a bit of simple coding.
We already leveraged computer power by moving from paper spreadsheets to digital spreadsheets, but they are limited. One thing I’ve seen over and over again — and occasionally been guilty of myself — is spreadsheet abuse. That is, using a spreadsheet program to do something I probably ought to write a program to do. For those times that you want something quick but want something more than a spreadsheet, you should check out Jupyter Notebooks. The system is most commonly associated with Python, but it isn’t Python-specific. There are over 100 languages supported — many community-developed. You can even install a C++ interpreter backend for it. Because of the client/server architecture, it is very simple to share notebooks with other users.
You can — in theory — use Jupyter for anything you could use Python for. In practice, it seems to get a lot of workout with people analyzing large data sets, doing machine learning, and similar tasks.
The Good: Simple, Powerful, Extensible
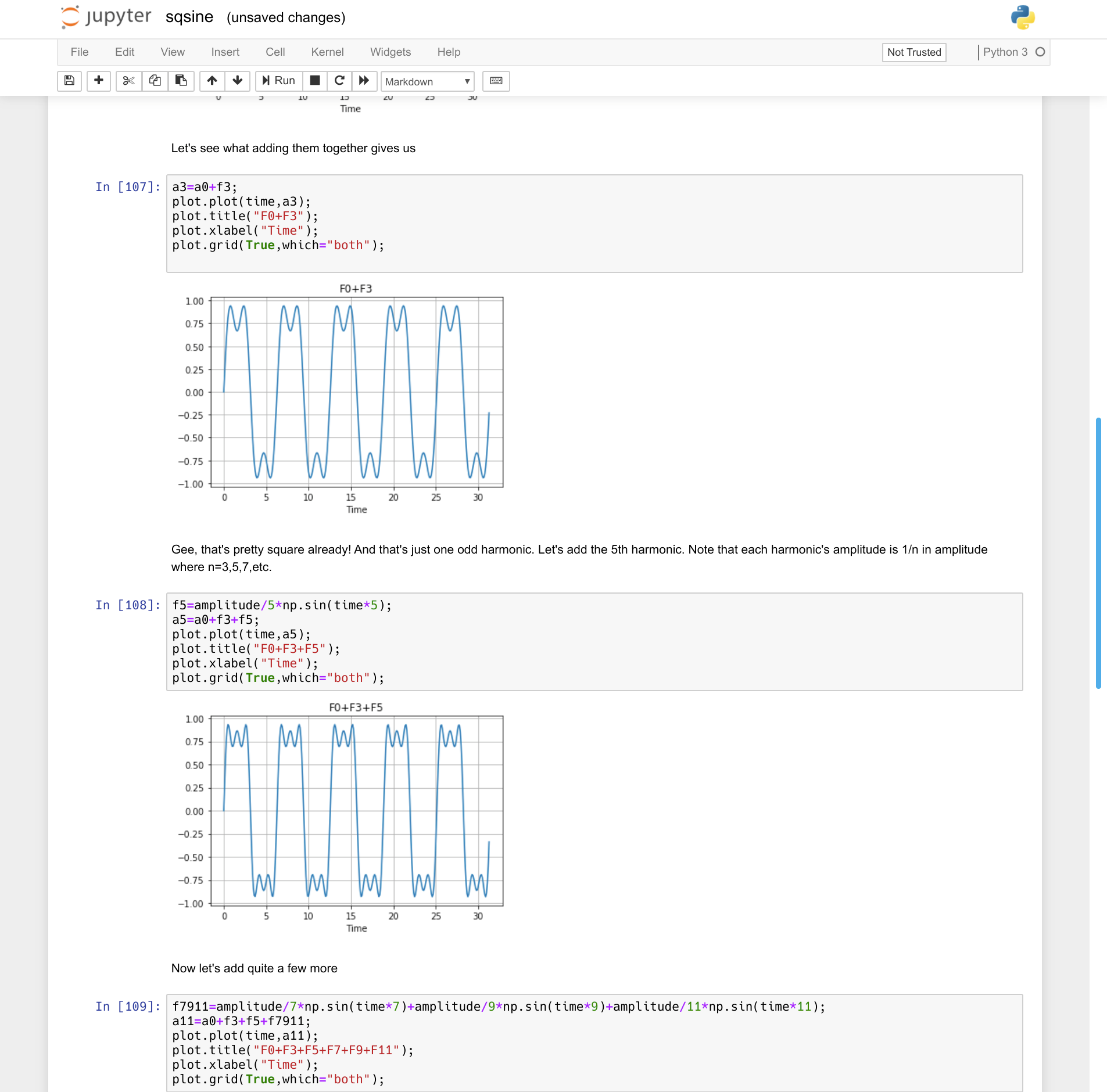
The idea is simple. Think of a Markdown-enabled web page that can connect to a backend (a kernel, in Jupyter-speak). The backend can run on your machine or remotely and will support some kind of language — often Python. The document has cells that line up vertically (like a single wide spreadsheet column). For example, here’s a simple notebook I created to explain how a bunch of sine waves add up to a square wave:
You can try it live in your browser or download it from GitHub. You can see that you can get “live” graphical output, along with text and other media. In fact, I’m not taking good advantage of the formatting, but you can do anything you can do with Markdown in the text cells.
The code is pretty standard Python. For example, here’s one of the cells:
a0=amplitude*np.sin(time); plot.plot(time,a0); plot.title("Fundamental"); plot.xlabel("Time"); plot.grid(True,which="both");
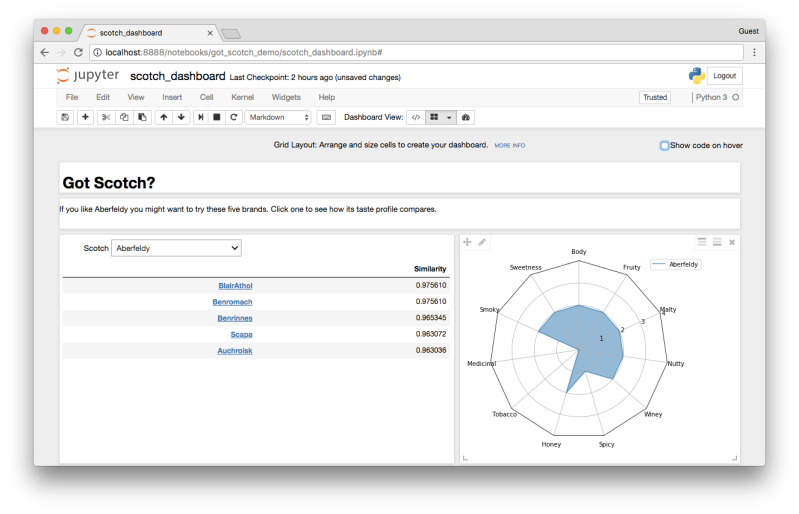
Further down in the document you’ll see that you can also deploy widgets. For example, using a slider to set parameters. We’ll come back to that topic in a bit. In addition to widgets, you can get extensions that let you layout cells in a grid. These are often used to create dashboards like the one below, for example. In fact, there are lots of extensions, for lots of different purposes.
The Bad: Support for Non-Python Languages
Non-Python languages are tricky to use with Jupyter. I tried using the C++ interpreter and found it a bit hard to get going. Some of that is because C++ isn’t happy with being run incrementally — Redefining things, for example, makes it unhappy. If you want C++ or Fortran or any of the other myriad options, they may or may not work well. They may or may not be able to use libraries that a lot of Python notebooks will employ. Don’t get me wrong. I haven’t found any that don’t work at all, but sometimes it is inconvenient or difficult compared to using the Python kernel.
The other thing that strikes me as odd is that the tasks notebooks seem best for is not always what they are most used for. If you think about it, the notebooks are really an exercise in literate programming. However, it seems to me that most of the notebooks are just sent around as quick web applications. You can share a static image of a page, of course. You can also share read-only versions. GitHub, for example, will render a notebook on display. There’s also Binder which will let you share an interactive version.
Joel Bennet does what he calls literate devops — which is similar to literate programming using Jupyter and — of all things — Powershell in the video below.
Jupyter is not magic. It facilitates rapidly building little Python applications that have a very particular web interface. There are probably projects it isn’t suitable for. Not every job requires a hammer. You can save yourself some grief, though, by doing a little research on best practices before you start anything substantial. But you should constantly be asking yourself if any tool is the right tool for a given job and not just using the same thing for everything.
The Ugly: Python Package Management
I find it taxing that the system relies on Python. I don’t have much against the language itself (although my personal preference is for whitespace to not be meaningful). However, ensuring Python has everything it needs for a given notebook tends to be super painful. If you plan on distributing, this becomes another layer of issues in ensuring everyone has the right packages. On Binder, you can provide a requirements.txt file that tells it what things you need to import, so that’s workable but an extra step.
The bulletproof way to install the program locally is with Anaconda which — of course — creates a totally different Python environment than your normal Python environment. Yes, I know about virtualenv. And pip. Of course, my Linux system has a package manager, too, and it has versions of Jupyter and all the Python libraries. But everyone wants their own package manager to rule my system and I have no idea what to do about your system.
Once you get it installed, it is fine. And if you get it working on Binder, you should be good since it builds each user a new Docker container. However, if you really plan on distributing complex notebooks, the installation across multiple platforms and Python versions could pose a risk.
Widgets Make It Interactive
If you look at the last two code cells of my example document (from above), you’ll see that I use a slider widget to let you interactively adjust the equations and the graph. That’s just one of the various widgets available.
If you aren’t picky, the system will build widgets for a function for you. You don’t always get the control of things like ranges and steps, but for many functions, you can get a reasonable UI by just making a simple call to interact, or including it with the function:
@interact(x=True, y=1.0)
def g(x, y):
return (x, y)
That will produce a checkbox for x and a slider for y. You’ll get default values, but in many cases, that’ll be acceptable.
The Latest
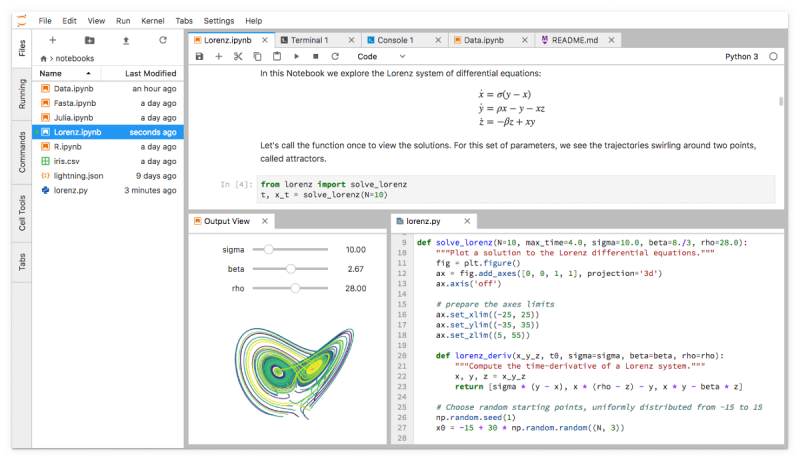
I’ve been talking about traditional notebooks, but the next generation interface rolled out last year. Known as JupyterLab, it allows you to use other tools like editors in a tabbed-interface. Binder supports the new interface if you want to give it a quick spin.
You can continue working with traditional notebooks using the new interface, so we expect to see increased adoption of JupyterLab over time.
So…
Should you use Jupyter? That’s like asking if you should use a saw. If you are cutting wood, yes! If you are trying to join two pieces of plastic, no. Jupyter definitely fits a niche — and a niche that many of us writing math- and data-intensive software work within. The fact that you can distribute it easily and even interface with hardware makes it attractive for projects where you want something quick but powerful.
Although some of the languages other than Python are second-class citizens, there are many choices and you can work around any limitations. So even if you aren’t a Python guru, you’ll still want to add this power notebook system to your toolbox.















If you don’t need graphing and/or embedding running codes, plain old markdown (or restructuredText) can be converted into html, pdf, odt and epub with pandoc.
I also like using Sphinx (with restructuredText) for writing manuals and tutorials. Even those that are not code related.
+1 for Pandoc
Every single document I produce at work and home begins life as a Markdown file only to be converted to whatever end format is needed at the moment.
Yeah but the point of using Jupyter is exactly needing to do those things. Jupyter is a competition/replacement to Matlab, not Markdown.
Not quite. Python with Scipy, numpy and Matplotlib (and other scientific/plotting/CV/AI libraries) is the replacement for MatLab. Jupyter provides a great front end to Python+libraries mentioned above that is easy to use and allows in my opinion much more flexibility than MatLab ever could. If I were doing research, Python + Jupyter + scientific libraries would be my first recourse.
However since I mostly create documents/tutorial/notes for students (I’m a teacher) I very much prefer using Pandoc and Sphinx.
“The idea is simple. Think of a Markdown-enabled web page that can connect to a backend (a kernel, in Jupyter-speak).”
Nice thing about JavaScript is the “back-end” is right there.
So THAT’S the nice thing about Javascript! I knew it’d be something.
I find myself staying away from object oriented. It seems the purpose is to protect the code from developers that don’t know exactly how to change the code. But if you’re a single man operation, you pay for it by having to a hundred places just to gain access to a variable.
The benefit is good on larger teams, or if your code gets so big so that you forget. I recently created a particle simulator (now on YouTube) and ripped out most of the object oriented stuff because editing to make changes was taking too long, and the compiled code was getting inefficient.
org-mode
Sorry but org-mode is not even close to what Jupyter can do. I love org-mode but for doing serious calculations there is just no contest between the two.
Guys, this is Jupyter is *not about writing documentation* (even though it can be used for that to some degree)!
So comparisons with Markdown or org-mode or “Markdown enabled webpage” are totally off the mark. Think more Matlab than Markdown.
Jupyter is first and foremost an environment to interactively perform numerical computations and to explore/visualize data, everything else is secondary to that. Also alternative languages than Python are supported quite well. I am only not quite sure why the author of the article picked exactly C++ to test it on – something that nobody uses with Jupyter. C++ is not exactly an interactive language and the support exists more as a curiosity than something usable. Try Julia, R, SPSS or even Matlab – you can even use all of them in the same notebook if you have the necessary software.
Re distributing notebooks – there are some best practices for it, including how to manage dependencies. For ex:
https://dzone.com/articles/pushing-jupyter-notebooks-to-production
No need to reinvent the wheel.
Re org-mode:
Org-mode can do some of the things but its support for anything beyond displaying a simple non-interactive plot is abysmal (well, that’s more Emacs’ fault than Org’s – Emacs wasn’t quite designed with such use in mind). EIN is a lot better but not maintained anymore and doesn’t handle any complex output all that well neither (certainly no embedded plots directly in Emacs).
What calculations or visualizations can I do with Jupyter that I can’t do with ‘normal’ Python? None?. As halherta says above, it is Python (and associated libraries) that is providing the alternative to matlab, Jupyter is just a way of running Python (or other languages). Very useful perhaps (especially for documentation/sharing/organizing), but ‘Jupyter is a replacement for Matlab’ is not really accurate IMHO.
IMO that’s wrong.
Yes, SciPy, Numpy, Pandas, &c provide the libraries that compete with MATLAB, but no one (or very few) programs MATLAB in a terminal or text editor. It;s a bit like saying ‘flour, sugar, eggs, butter, vanilla, and baking powder make cupcakes, not the pan you bake them in”. When people are decide to use Jupyter notebooks it’s not because they can’t effectively use the python libraries that give it power. They use it because they want or in a few cases need the iterative code execution and cell separation Jupyter gives them.
You’re both missing the forest for the trees.
Scipy, Numpy, Pandas + Python is equivalent to MatLab’s programming language (terminal + text editor edition….yes people do that….not every one will use the GUI…besides the MatLab GUI last I checked basically brought together a Matlab REPL, text editor, debugger and a few other nice graphical tools).
Jupyter + Python + libs is similar to Matlab’s programming Language + GUI interface
Jupyter is a nice front end for data science and those that are too scared of the command line or find it to be a waste of time. But it needs Python or some other language to make it useful.
If one only needs to create notes, especially if these notes do not rely on dynamic graphs or embedded live coding examples, then Jupyter may not be the best tool for the job. In this case have a look at Pandoc+Markdown / RestructuredText. If you want to generate a manual, tutorial e.t.c. also have a look at Sphinx.
I’d go the other way and say it has nothing at all to do with Python, and all the features come out of having a sharable REPL (read-eval-print loop).
Traditional use of a REPL, you type code in to try the code out, and at the end of the session it gets thrown away; if you want to keep something, you use an editor to create a file and paste it in. Here, the REPL itself is a savable, shareable document. It even groups a small number of related lines together in a chunk, kindof like a method or function, but that doesn’t require any boilerplate.
The only reason that Python has anything to do with it is that Python is the preferred simple language for people who aren’t primary programmers; the BASIC of the modern age. If you used Perl or Ruby instead, the value would still all come from having a shareable REPL.
“Spreadsheet abuse”, no.
https://www.youtube.com/watch?v=UBX2QQHlQ_I
I use this personal wiki http://zim-wiki.org/
Emacs/Orgmode/Babel may look a bit robust and chunky sometimes but if you want to preserve the information (this should be your primary goal!) and the (current) presentation is not the most important aspect, your’re on the safe side with it.
Emacs/Orgmode/Babel is absolutely polyglot and you’ll find help nearly everywhere.
I just want something easy/simple to put together a program to remote control an old Toshiba projector via RS232. The commands are text. Click a button, send text to the selected COM port, wait for the acknowledge. One GUI button per command. Preferably in a tabbed interface for grouping buttons by function.
PyQt and Python3.
yes, definitely, and if you like wysiwyg approach, use the QtDesigner/QtCreator program. For something very simple, you might also consider tkinter (built into python) but this is quite limited in my opinion.
There is something wrong with my jargon today. I can’t figure out the title.
Anyway, Jupyter is the cat’s pajamas. https://nbviewer.jupyter.org/github/ARMWorks/IPNB/blob/master/sampleIPNB.ipynb
The title is a reference to the only listenable song by a boring band called Train.
That is a lie, there are none.
This looks easier than I expected. My notes are usually plain text files, sometimes google docs, and supporting files tossed in the same directory. Thinking about it now I should probably just switch to markdown, that would cover both plain text and rich text and still be easily editable. So thanks!
Re dependencies: check out https://github.com/jrjohansson/version_information for a good way to help communicate the dependencies when distributing your notebook.
I use jupyter Notebook for technical conceptual calculations, including minimal documentation for later, prototyping image recognition, scientific calculations, condensing/comparing simulation results, and funky interactive presentations. It is a helpfull tool. Started using it as a alternative to mathcad.
I was just thinking of Mathcad as well. My ‘problem’ is that I’m a software developer and can definitely use an ipad-like device that lets me do simple calculations, as wel as test some basic algorithms, and maybe generate a precalculated table, or format a binary file in decimal representation, or quickly build a SQL schema, import some data and execute some queries. And all of that visually and intuitively, because I don’t want to RTFM. ;)
Jupyter stands for “JUlia,” “PYthon,” and “R.” Supposedly you can mix and match all three. Jupyter can also run Scilab and Octave code (among so many other languages), provided the appropriate run kernels have been installed. I think you can also run MATLAB from inside Jupyter, but I am not 100% certain on this. In recent years, the Scilab and Octave teams put a lot of effort in interoperability with Python. Take a look here, for example: https://forge.scilab.org/index.php/p/sciscipy/page/Tutorial/. And here:https://news.scilab.io/news97/. This could be really helpful because Scilab has something similar to MATLAB’s Simulink, called XCOS, which the Python-based tools lack. So, it should be possible to set up a simulation model in Scilab using Scilab’s excellent tools and then run it from inside Python. (Disclaimer: I am not up to date with XCOS, but I do know that Octave has also adopted Scilab’s XCOS.) You may want to take a look here for more information on how to use Scilab XCOS models from inside Python: https://stackoverflow.com/questions/24506618/how-to-use-model-from-scilab-xcos-in-my-program .
Jupyter is not based on plain Python, but on IPython. That makes a big difference. For example, in IPython you have the so-called magics. You can declare a cell (section of code) as Javascript, Scilab, Octave,…, and just run it inside the current Jupyter session. (That is different than passing data between such programs and Python, which can also be done from Scilab and Octave.)
Jupyter also makes Python’s parallel processing a lot easier to use by providing a graphical tool to manage so-called IPython clusters (essentially parallel engines). And with the %%cython magic, you can run in-line Cython code inside the same Jupyter environment. (Cython is COMPILED Python; it runs as fast as C and invokes the C compiler, but you have to prepare the Python data structures that will be passed to Cython). Magic cells are a feature of IPython and not specific to Jupyter, but Jupyter makes it easier to use them. The Jupyter notebook started as an IPython project (and if fact was called “IPython Notebook”), but about three years ago was separated from IPython and became usable with other languages/packages.
Where I find Jupyter problematic is with the use of pdb, Python’s debugger, which is similar to gdb. However, there is another IDE tool, called Spyder, which is very MATLAB-ish, which works well with pdb. Spyder is also included with Anaconda and there are efforts to make Spyder and Jupyter interoperable, but I am not up to date on this either. Spyder can be installed as a stand-alone application as well and works with plain Python but also IPython. Spyder’s web site is https://www.spyder-ide.org/ . As I said, it is very MATLAB-like.
Personally I would like to see TeX/LaTeX support integrated, not just markdown. And maybe usability with PyCharm? I really like PyCharm as a nice Python development environment, especially with the nice friendly management of packages and virtual environment automatically spun up for your project.
Personally I would like to see TeX/LaTeX support integrated, not just markdown. And maybe usability with PyCharm? I really like PyCharm as a nice Python development environment, especially with the nice friendly management of packages and virtual environment automatically spun up for your project.
Google Colaboratory is the most convenient way to use Jupyter Notebook, you should have mentioned it.
http://colab.research.google.com
I’ve been using tiddlywiki. It is just and html file. You need to invest some minutes to see how to allow ‘saving’ itself with today’s browsers security settings. Highly recommended