In order to understand something, it helps to observe it up close and study its inner workings. This is no less true for the brain, whether it is the brain of a mouse, that of a whale, or the squishy brain inside our own skulls. It defines after all us as a person; containing our personality and all our desires and dreams. There are also many injuries, disorders and illnesses that affect the brain, many of which we understand as poorly as the basics of how memories are stored and thoughts are formed. Much of this is due to how complicated the brain is to study in a controlled fashion.
Recently a breakthrough was made in the form of a detailed map of the cells and synapses in a segment of a human brain sample. This collaboration between Harvard and Google resulted in the most detailed look at human brain tissue so far, contained in a mere 1.4 petabytes of data. Far from a full brain map, this particular effort involved only a cubic millimeter of the human temporal cortex, containing 57,000 cells, 230 millimeters of blood vessels and 150 million synapses.
Ultimately the goal is to create a full map of a human brain like this, with each synapse and other structures detailed. If we can pull it off, the implications could be mind-bending.
Neurogenesis
In the beginning, there exist just a few newly formed neurons, gradually specializing from more generic cells in the developing embryo into the variety of cells that make up the human brain. Studying the development of this early state into the child and adult connectomes (that is, the map of neural connections in the brain) has provided us with many clues already. The term ‘connectome’ was originally coined in 2005, following the pattern of ‘genome’ at a time when researchers were busy sequencing the full human genome.
The idea is that in order to understand the brain, we must understand the network and not merely the individual neurons or functioning of individual synapses. This is similar to how our understanding of a genome relies on observing how DNA is transcribed into functional building blocks; components which ultimately result in anything from as simple as a single-celled eukaryote, to a functioning human body, or any other lifeform. By observing the way the genome is used in this context, we gain understanding of how both the genome and the cell or body it’s part of fit together.
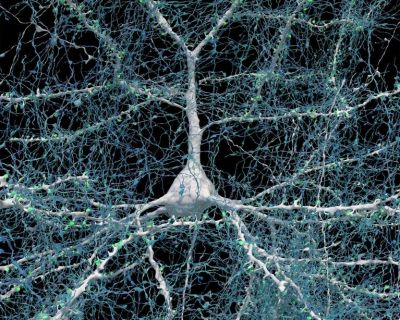
In the case of a connectome, we have been rather limited in our observation options, as brains do not take that kindly to being studied with e.g. a scanning electron microscope (SEM) while still inside a living body. Thus it is that most studies involve the use of functional magnetic resonance imaging (fMRI), which images brain activity over time. A recent study by Qiongling Li and colleagues in Cell Reports titled “Development of segregation and integration of functional connectomes during the first 1,000 days” used fMRI sets of scans on 665 infants to track the development of their functional connectivity (FC) up to the age of three years, which is when the brain’s structure has largely matured.
Using this dataset, it is possible to mark the segregation of the developing brain into distinct hubs, which form the various cortices and so on, each with distinct functionality. Local connectivity can be tracked using this method, as well as global connectivity between the separate regions, but this does not tell us how exactly the neurons are wired up, nor does it allow us to take a close look at the developing connectome, or the final result in an adult.
Adult Connectome
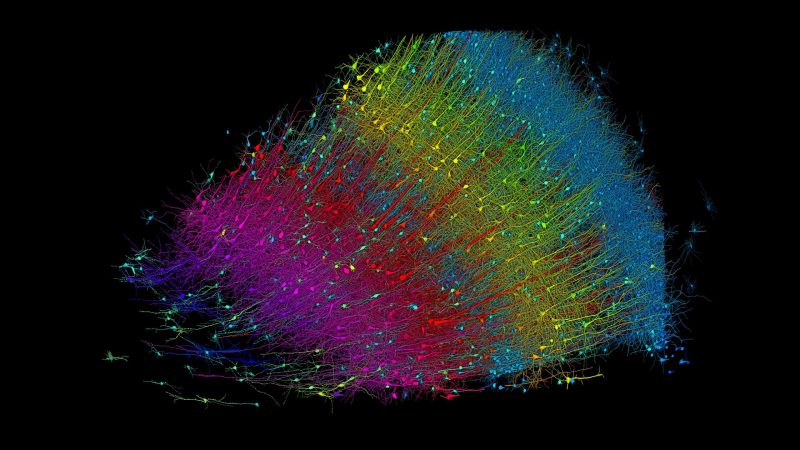
The brain tissue sample that was analyzed in the aforementioned collaboration study by Google Research and Harvard’s Lichtman Lab came from a 45-year old woman who underwent epilepsy-related surgery (surgical resection of an epileptic focus in the left medial temporal lobe). The study results are described in the research article (paywalled, no prepublication copy found) by Alexander Shapson-Coe and colleagues, as published in Science with the title “A petavoxel fragment of human cerebral cortex reconstructed at nanoscale resolution”. Detailed information on the materials and methods can be found in the (freely available) supplemental material PDF.
This 1 mm3 sample was stained first with tetroxide-thiocarbohydrazide (TCH)-osmium, washed and then stained with 2% uranyl acetate prior to being fixed with epoxy. From this 5019 sections were cut, ranging in thickness from 30 to 33 nm, albeit with issues such as layers breaking up, blunting of the used knife and alignment issues after a knife replacement. This is presumed to have led to some loss of the tissue.
These slices were subsequently imaged with a SEM, leaving the researchers with the task to align and match the features in each layer. This process was handled mostly by machine vision and algorithms such as OpenCV’s SimpleBlobDetector, as well as proprietary Google algorithms. Convolutional neural networks (CNN) to perform tissue and neurite classification as well as myeloid body and data irregularity detection. Ultimately, after a lot more processing, the result was a viewable 3D data set with the individual elements like neurons, glial cells, blood vessels and synapses.
These datasets are publicly available, using the Google Neuroglancer web-based tool for interactive viewing. As noted by the researchers, the merging of the layers is far from perfect, and a lot more proofreading by human reviewers will be needed. Even so, it provides a fascinating glimpse at a world that’s normally impossible for us to observe so clearly.
Mapping Implications
Even in this small tissue sample, the researchers made a number of discoveries that raised questions about normal neural development. In particular, they noticed quaint characteristics, such as a few neurons that had established over 50 connections with each other, as well as knotted dendrites. These were unlike what had been documented before, which raises many questions about whether these might be indicative of some kind of developmental issue or not.
Within this small dataset it’s possible that these are features associated with epilepsy – since this sample was obtained from a person’s brain who suffered from that condition – but without obtaining more data to compare with it will be exceedingly difficult to draw any firm conclusions. This then leaves us for now with both a very exciting outlook and the gloomy realization of how hard it will be expand on this effort.
As the level of detail from this one experiment’s results show, there is a lot that we can likely learn about the brain’s structure and functioning, especially once we manage to characterize even more brain tissue, allowing us to compare and contrast between healthy individuals and those we suffered from a variety of conditions in life. Due to the scanning method used, the tissue has to be either in the form of a biopsy (as in this case), or from a post-mortem donation, which adds its own set of challenges.
The effort required in preparing just this one 1 mm3 sample, the enormous dataset it generated, and the still imperfect post-scan processing and reassembly process poses significant hurdles. Considering that the average adult human brain has a volume of around 1200 cm3 (1,200,000 mm3), the human genome sequencing project suddenly looks like an easy afternoon challenge.
That said, much like the human genome project we will likely figure out ways to optimize the brain mapping project. Once we do, we can expect a flurry of new insights on aspects about the human brain that have puzzled us for centuries, which would absolutely make it worth the trouble.














I’m reminded of the cruel uncle: When a child asked how a magnetic compass worked, he answered “Why don’t you take it apart and find out?”
Excellent analogy, actually. Wow.
For something disturbing—
https://en.m.wikipedia.org/wiki/Cortical_homunculus
Okay so assuming an average male brain volume of 1260cc, that equals around 1.8 zettabytes of data. And then you have to start crunching it to actually figure out what all the circuits do. And then you have to realize that synapses aren’t like regular electrical circuits, there are many different neurotransmitters, and there are almost certainly other mechanics that affect the whole system including from other parts of the body.
Don’t expect the source code to human consciousness anytime soon is all I’m saying. Certain people have a very impressive level of knowledge about the brain—so this isn’t a slight towards them—but when compared to a complete understanding, the best of us only have a hilariously small portion of it.
Maybe start with a simpler brain, like that of a mouse. I seriously cant understand why governments aren’t piling truck loads of cash into such a project …. that said, the N.Koreans have probs done it already.
Why even spring for a mouse brain when they could pick yours?
I was going to offer a specific part of my brain that I’m not using any more.
They are, ChatGPT is a fully functioning vice president.
Think smaller, start with Etruscan shrew. It has a brain mass of about 60 mg. Where as a typical lab rat has a brain with a mass of about 2 grams on average.
This message has been fully approved of by Pinky and the Brain!!
I vaguely recall seeing an entire brain mapped in such a way. Maybe it was the nematode or something. Some tiny critter with not that many neurons. Certainly at this time even the humble fruit fly is out of the question.
This is true – brain of fruit fly is massively compacted for weight and size efficiency. It’s amazing how intelligent fruit flies are.
They’re pretty sophisticated.
https://www.noemamag.com/the-surprisingly-sophisticated-mind-of-an-insect/
And I think this data only captures the connections between neurons but not the synapse activation thresholds. The activation thresholds are partly an electrochemical phenomenon so the information about them is likely lost very soon after the tissue is removed from a living being.
Quantum effects need not apply.
https://physicsworld.com/a/do-quantum-effects-play-a-role-in-consciousness/
Very true. But it’s fun to imagine if we did have it all. My first though reading this was imagining.. if they actually had ALL the information in a brain. Then wrote an “emulator” to run it. With the best computers available.. how slow would that be? I imagine it running for the remaining lifetime of the universe.. the poor mind gets to experience a few seconds of existence.
They did that. The answer is 42.
.
There’s a whole series of documentary books about.
Gee, maybe if Google would use all that memory they’ve been using to store what they’ve accumulated about each of us., and instead use that memory to store this type of data, they’d accomplish something good for humanity.
Risky, allowing a single company to know so much. Cyberpunk/deus ex outcome would likely ensue. I would hate to be in a world where one can buy knowledge than read/study on ones own.
Are you for real? Did you just say that if Google had extensive knowledge about the human brain it would be good for humanity?
Wow… OK
I was hoping to sidetrack Google from their current trajectory.
B^)
Luckily they are already all over the place, starting and dropping projects in such a chaotic manner that sometimes they cancel a project even before it started but then still go through the process of introducing it and advertsing it while anybody trying to use it finds it doesn’t exists ‘anymore’.
Hmmm, so when the technology catches up we could, in theory, take a dead persons brain, slice and dice it, map it out and then use the data to simulate the brain in software like running an NES emulator on a Pi?
To paraphrase Douglas Hofstadter: How do you know this has not already happened?
More precisely: How can you tell it has not already happened to *you*?
Remember, reality is an illusion, the universe is a hologram, buy gold, BYYYEEE!
This could explain a lot of my life decisions.
Well if it has I want to have a word with the SysAdmin.
That might result in a
Kill -9
Or
Sleep()
Not quite the same thing but this is still a cool braon map i ran across this morning
https://www.reddit.com/r/coolguides/s/ko15ydDxN4
It could be curing of mental diseases or world peace.
But it is going to be ads directly into the brain.
At this point the entire world rotates around mental disease and I’m not sure if we could handle the chaos of curing such.
We’d have to reinvent EVERYTHING and replace all of which current societies are based on.