OSM stands for Oligonucleotide Synthesizer designed for use in Microgravity, meaning that it’s a device that makes arbitrary DNA strands (of moderate length) in space. Cool eh? I’ve been working on this project for the last eight months with a wonderful team of fellow hackers as part of the Stanford Student Space Initiative, and I’d like to share what we’re doing, what we’ve already done, and where we’re going.
Why space? Well, first of all, space is cool. But more seriously, access to arbitrary DNA in space could accelerate research in a plethora of fields, and the ability to genetically engineer bacteria to produce substances (say on a martian colony) could mean the difference between death and a life-saving shot. In short, it’s hard to predict the exact DNA one might need for research or practical use before hand.
First, as Hackaday tends to be a little light on biology terminology, we need to get a little vocabulary out of the way to grease the ways of communication. If you have a Ph.D. in synthetic biology, you might want to skip this section. Otherwise, here are five quick terms that will make your brain bigger so stay with me!
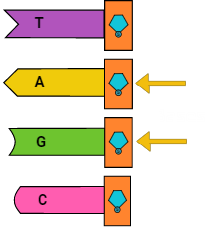
Deoxyribonucleic acid (DNA)
Since DNA is the focus of this project, it’s paramount that one understand DNA before proceeding. But, since most have seen DNA in high school biology courses I’ll keep it short and simple.
First, DNA is a polymer composed of nucleotides. Nucleotides are the building blocks of DNA, and consist of one of four nucleobases — cytosine (C), guanine (G), adenine (A), or thymine (T) — a sugar called deoxyribose, and a phosphate group. Different nucleotides can intermolecularly bond to corresponding nucleotides (A to T; C to G) on a different strand of DNA. If the bases on a strand of DNA are the complements of the bases on another strand, the two polymers can bind and form double-stranded DNA. And, one more note: DNA has two different ends: a three prime (3′) end and a five prime end (5′).
Oligonucleotide (Ah-li-go-new-klee-o-tide)
Oligonucleotides are short DNA or RNA molecules and are super useful in general research, genetic testing, and bioengineering. The length of an oligo (short for oligonucleotide) is usually denoted as 30-mer or d30 in the case of an oligo with 30 bases. How short does a strand of DNA have to be to be considered an oligo? Well, back when we were… let’s say not-so-good at DNA synthesis, oligonucleotides were firmly in the 2-50 range. However, with the advent of phosphoramidite (an inorganic DNA synthesis method), oligonucleotides can range from 2-1000+ bases in length.
Homopolymer
An oligonucleotide comprised of only one type of base (eg AAAAAA as opposed to AGTCTG) is called a homopolymer.
Terminal Deoxynucleotidyl Transferase (TdT)
Terminal Deoxynucleotidyl Transferase, commonly abbreviated to TdT, is a polymerase-like enzyme that can add arbitrary nucleotides to the 3′ end of a DNA strand when certain conditions are met. TdT can append all four nucleotides but shows a preference for guanine (G) and cytosine (C). TdT can be found naturally in immature, pre-B, and pre-T lymphoid cells where it performs genetic diversification for our immune systems by adding bases onto the 3′ (three prime) end of our DNA.
SYBR Green I
SYBR (pronounced like cyber) is a molecule that binds to DNA forming a DNA-dye-complex that absorbs blue light and emits green light. Moreover, SYBR preferentially binds to double-stranded DNA but will bind single-stranded DNA to a lesser extent.
</vocab>
Goals of Making DNA in Space
Now that we’re speaking the same language, let’s take another look at the goal of making arbitrary DNA in space enzymatically. What exactly does that mean, and why space? Okay, let’s take this word by word:
- Arbitrary: Our process shouldn’t simply make copies of DNA (like PCR) but should instead create any sequence (eg. AGCTATC) that one can type into a computer (and can physically exist).
- DNA: I think y’all got this one
- In space: Obvious meaning: we want our final device to work in space, which imposes all sorts of physical and chemical restrictions.
- Enzymatically: We want to use a new method of DNA synthesis using enzymes instead of the dangerous and resource intensive phosphoramidite method.
Adding the ability to manufacture custom DNA in microgravity may mean a huge leap forward in our ability to survive away from Earth. OSM serves as a biohacker’s toolkit for unprdictable problems we will face in new environments when we support life on and around other planets.
In summary, we want to create definable oligonucleotides in the demanding environment of space with a new enzyme-based method. They say set your goals high, right?
Minimally Viable Product (MVP)
Okay, maybe not that high. At least, not initially. Rather than jump in all or nothing, we’ve chosen to take an iterative approach. Now, we are working on launching a “Minimally Viable Product” for a few reasons:
- To validate our work thus far
- To become comfortable with space ready design
- To get real space data
- To gain credibility for future launches
MVP Process
As this is our Minimally Viable Product, we’re taking a small bite out of our overall goal. As such, our MVP will synthesize a homopolymer on the end of an existing oligo rather than create completely arbitrary DNA. We won’t have complete control over its sequence (all in good time) but we will demonstrate a proof of concept of some of our work.
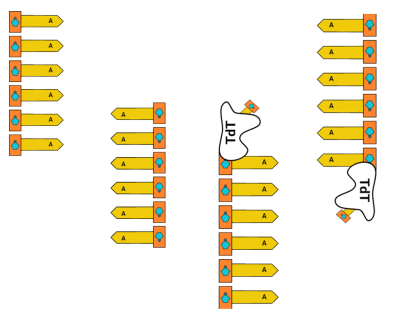 First, we’ll start with homopolymer adenine (A) strands of DNA in solution. Once in space, we’ll raise the temperature and allow TdT to append thymine (T) to the strands.
First, we’ll start with homopolymer adenine (A) strands of DNA in solution. Once in space, we’ll raise the temperature and allow TdT to append thymine (T) to the strands.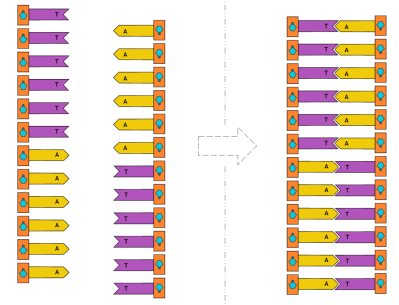 Because thymine (T) and adenine (A) are complementary, the single-stranded DNA should form double-stranded DNA.
Because thymine (T) and adenine (A) are complementary, the single-stranded DNA should form double-stranded DNA.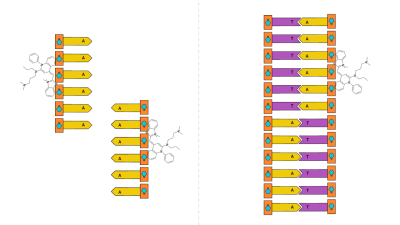 Then, we’ll use SYBR Green I to detect if there is double-stranded DNA in solution, to determine if our experiment worked. This works because SYBR preferentially binds to double-stranded DNA, and only fluoresces if bound to DNA. This verification step is incredibly important because we’ll be unmanned, and science done without verification isn’t really science.
Then, we’ll use SYBR Green I to detect if there is double-stranded DNA in solution, to determine if our experiment worked. This works because SYBR preferentially binds to double-stranded DNA, and only fluoresces if bound to DNA. This verification step is incredibly important because we’ll be unmanned, and science done without verification isn’t really science.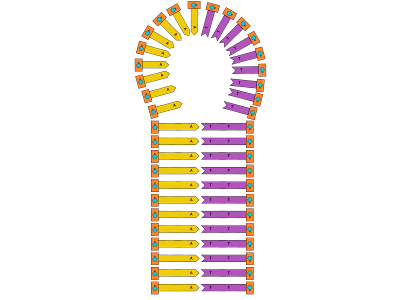 Additionally, hairpin DNA or more formally, Stem-loop intramolecular base pairing, is something that has been brought up as a possible issue with this design. If the DNA binds with itself, it can’t bind with other strands. Sounds like a problem, right? But look closely: if the DNA binds with itself. Even if we form hairpins rather than “proper” double-stranded DNA, we’ll still have double-stranded DNA, which we can detect.
Additionally, hairpin DNA or more formally, Stem-loop intramolecular base pairing, is something that has been brought up as a possible issue with this design. If the DNA binds with itself, it can’t bind with other strands. Sounds like a problem, right? But look closely: if the DNA binds with itself. Even if we form hairpins rather than “proper” double-stranded DNA, we’ll still have double-stranded DNA, which we can detect.
In review, we’re using an enzyme whose purpose is genetic diversification with SYBR Green I, a molecule commonly used for staining gels used in electrophoresis to synthesize DNA in space.
Now, while this sounds simple in practice, synthetic biology always sounds simple. However, in my biology experience, nothing is ever simple. All sorts of data are needed so we can be confident our device will work. What we’ve done so far:
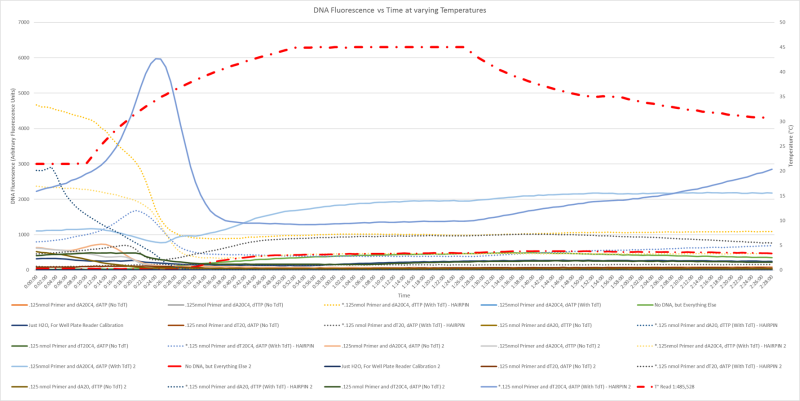
- Confirmed that we can synthesize homopolymers with TdT effectively and reliably with all nucleotides
- Determined long-term compatibility of our reagents (they might sit together for 2+ months)
- Explored temperature dependence of SYBR Green I
- Determined the levels at which we can detect the difference between single-stranded and double-stranded DNA with SYBR Green I
MVP Design

Working with enzymes in space is hard. TdT specifically requires controlled temperatures, buffer solutions, and a non-metal container, all sustained for more than a month in space. Moreover, we need a verification system on board, which means we’ll need fragile filters and visual access to our samples.
Our initial design features a 50μL PDMS (a clear bio-safe silicon-based organic polymer) reaction chamber surrounded by an aluminium heat equalizer heated by Peltier modules. The Peltier modules currently transfer/absorb their heat to/from a generic heatsink rather than something more application specific, as we don’t know what our launch conditions may be. Moreover, a photodiode, filter and LED hug the PDMS reaction chamber, allowing for fluorescence tests to verify experimental success.
Beyond MVP
As our MVP is just a stepping stone to our ultimate goal, we’ve been working on all sorts of other things. For instance, we’re working on an electrowetting on dielectric device (see right) as a stand-in for conventional microfluidic devices commonly used in space.

We’ve also explored pyrosequencing and synthesis on magnetic beads. Additionally, we talk about DNA so much that we made bare bones DNA visualization software to facilitate conversation. (The DNA pictures in this article were generated by this software.)
However, as far as we’ve come, we’re still flushing out our design and would love any suggestions you might have. Specifically, we’re having problems with finding PCB manufacturers that have sub 1mil minimum trace spacing. Tips, advice and healthy conversation on this are all welcomed in the comments below.













does it ever give you pause, that your project is treading dangerously close to Seed Theory territory https://en.wikipedia.org/wiki/Panspermia
I mean, that’s sort of the idea here isn’t it? If we’re going to colonize other planets, we need an ecosystem that can actually survive it.
Given any thought to the errant or malicious DNA that might be generated in this random process? And where it might eventually end up?
It doesn’t appear to be random, but arbitrary; you input the material, and what you want to produce. The arbitrary part seems to just imply that it’s meant to be general use, with no one specific type of DNA in mind.
That’s my understanding anyway, I could be wrong.
Exactly, we aren’t making any choices about what DNA to produce, that’ll be up to the end user. As of right now, we’re working with homopolymers and short arbitrary sequences
We have, and we’ve come to a few conclusions: 1. Since the end user can choose their sequence, it’s up to them to not make malicious DNA 2. Even if we wanted to put in safety controls, there is little way for us to prevent the creation of malicious DNA as we would need to know all malicious DNA sequences and we would also need to handicap our protocol, both of which are impractical/infeasible
Anybody can just order “malicious DNA” from any cheap and convenient oligosynthesis lab online – it’s nothing new.
Noodle suffering from being terminally baked. We weren’t made of DNA when I did biology in the 60s. :)
For sub 1-mil traces, have you checked Epec (http://www.epectec.com/)? They do several special processes, though I don’t know if they do traces that small. Their sales & engineering support staff have been helpful and knowledgeable for us.
Too, bravo, HaD. Opening comments are two for three with “OMG your research uses a Sci-fi doomsday keyword – j00 willz killz us awllll!!!1!!”
“treading dangerously close” may have been better phrased “loosely resembling (after a few pints)”. Our spacecraft are clean but not sterile (easy ref – look at how careful NASA is being with Cassini right now); that output dwarfs the OP’s, which is dwarfed by orders of magnitude by the actual suspected carriers.
“errant or malicious DNA” “random process” – I think you meant “carefully set up and contrived process” and “stuff that is totally likely to be dangerous if this is a movie on the SyFy channel”
I think that it is either a nice space science student experiment, but without all they features they talk about, or they left out a lot from the science description. In either case I see a lot of unnecessary hype which is not typical for scientific setting. It is not even a synthetic biology problem, or even a problem at all because it is already solved. Arbitrary sequence oligos are the workhorse of molecular biology lab, you can get them for a few bucks. So why the hype? The problem with science I can see is that this will not produce arbitrary sequences in its current form. TdT will be adding nucleotides at random from a mixture of nucleotides. Therefore produced DNA will have random sequence and not arbitrary sequence chosen by you. So, let’s add a little spin and add only one nucleotide at a given time and the then cycle through them. Ok, it could work if the enzyme would add just a single nucleotide per cycle, but the problem is that we cannot control that. So it will be adding homopolymers of random length. Instead of nice ACGTA, you will have a random mixture of AAACCGGGGGGGTAAAAA, AACGGGTTTAAA, etc. Unless they have an idea how to solve the problem of adding one nucleotide at a time, though they do not mention that.
You’re completely right. Given only the things presented in this article, we don’t have much to offer as far as oligonucleotide synthesis here on earth. We’re working on multiple single specific nucleotide additions via the use of uv-labile blocking groups (2-Nitrobenzyl) as our next step which answers your concerns. As you said, “it could work if the enzyme would add just a single nucleotide per cycle.” I didn’t go into more detail about this for a few reasons: (1) The article was already lengthy, and (2) we are still gathering data and haven’t perfected our methods yet. All good stuff!
PS the hype is mostly due to our enzymatic method which provides a variety of benefits specific to use in space over the commonly use phosphoramidite methods used here on earth
Ok. Cool project for the student initiative. I just fretted over passing it as a general purpose oligo synthesizer in its current stage. But it is a great ultimate goal for the project.
Using TdT for DNA synthesis is a difficult (and at this point unsolved) problem, but conventional DNA synthesis uses some pretty flammable/toxic phosphoramidite organic chemistry which makes it difficult to put those synthesizers in new places. For example, anhydrous acetonitrile is used to flush the tubing between every step and has a flash point around 2 degrees Celsius, and is pretty toxic to breathe. The all means that right now phosphoramidite chemistry is pretty much a non-starter to deploy in places like space, so we have to try something totally different (TdT).
Will you also need to have a matching amplifier and sequencer so that you can scale up and validate the output before use?
Short answer: Possibly. For an amplifier, normal PCR with primers (possibly made by OSM) should be sufficient. Furthermore, our goal is to not need a sequencer. We hope to be accurate enough that one can trust the output of OSM to be sufficiently correct
It’s a nice idea. Oligosynthesis from commercial labs is cheap and convenient – but it’s easy to take that for granted, and you don’t have access to that in space. Oligos are the underrated, quiet workhorse of molecular biology – without custom oligosynthesis you don’t have PCR, you don’t have Sanger sequencing, and you don’t have all sorts of important stuff.
I wonder if the hardware can be made as a more general-purpose apparatus that could be used to perform multiple different jobs. You basically have a microscale reaction vessel, microfluidics, heating and temperature regulation, and electro-optical readout of the SYBR fluorescence signal. So what else could you do with the same hardware?
In particular, quantitative PCR (or ordinary PCR), or loop-mediated isothermal amplification, or Sanger sequencing all come to mind as potential possibilities. Somebody in the comments above mentioned sequencing – I believe they’re already using a nanopore MinION on the International Space Station, apparently it works in microgravity.
Are there any existing documents that say what would be unacceptable in terms of chemical hazards, say on an ISS experiment, and what is acceptable? I’m sure they’re capable of small-scale safe handling of chemical reagents – there has been much materials science and biology and chemistry done on the ISS for many years. I believe all the biology stuff is rigorously contained in a glovebox anyway.
The use of microfluidics and small reaction volumes reduces the chemical hazards, so that’s good. If TdT synthesis doesn’t turn out to be practical then I would fall back to proven phosphoramidite synthesis, and design the hardware accordingly. I’d rather take up the acetonitrile and what not rather than not have any access to oligos in space. It’s not that toxic, flammability is probably the main concern.
Michael – typo.
> An oligonucleotide *comprised of* only one type of base (eg AAAAAA as opposed to AGTCTG) is called a homopolymer.
Composed of, comprises. Source: http://www.quickanddirtytips.com/education/grammar/comprise-versus-compose
Expected more from a Stanford student!
By the way, for sub 1-mil traces, you could look at metal electrodes patterned on SiO2. You can go down to a few dozen microns almost trivially that way, and I’m sure a lab on campus has the facilities.
https://www.instructables.com/id/Digital-IC-Tester-for-Industries-and-Engineering-C/
https://in.linkedin.com/in/shubham-kumar-746b84108
https://hackaday.io/shubham2ece
Check it out.
This is really interesting ! Do you guys have a drawing of the reaction chamber / microfluidic setup. I would definitely like to talk to you guys a little more about this project. I have a question though, I feel like doing this same experiment in microfludics will have the same effect as microgravity.
In addition to that, depending on the altitude of your (small sat) I feel like you’re entire system might be affected by radiation, it might be hard to separate the effect of microgravity vs radiation. I design microfludics CAD tools as a part of my research and I worked on space systems engineering before, let me know if there’s anything I can help you with.
Great science but I can imagine some idiot producing radiation resistant lunar slime mold.