Hitting the electronic surplus shop is probably old hat to most of our readership. Somewhere, everyone’s got that little festering pile of hardware they’re definitely going to use some day. An old fax is one thing, but how would your partner feel if you took home an entire pallet-sized gene sequencing rig? Our friend [kaspar] sent along an interesting note that the folks at Swiss hackerspace Hackteria got their hands on an Illumina HiSeq 2000 last year (see funny “look what we got!” photo at top) and have generated a huge amount of open documentation about whats inside and how to use it.
Okay first off, what the heck is this machine anyway? The HiSeq is designed to automatically perform the sequencing step of Illumina’s proprietary multi step gene sequencing process (see manufacturer’s glossy for more), and to do so with minimal human intervention. That means that the unit contains a microfluidics system to manipulate samples, an extremely high performance optical scan system complete with controllable stage, imager, fluorescence modes, etc, and lots of other things this author isn’t sufficiently trained to guess at.
The folks at Hackteria have done a pretty thorough teardown of the system and produced block diagrams of most of its modules. They’ve also run some of the tools and recorded logs of what they were up to, including the serial commands sent to and from the machine to control certain subsystems. Of course a tool like this was meant to be driven by Illumina’s specific software, but unusually those are available and surprisingly usable which is how the aforementioned logs were captured. Right now it looks like Hackteria has put together tools to use the system as a fluorescent microscope.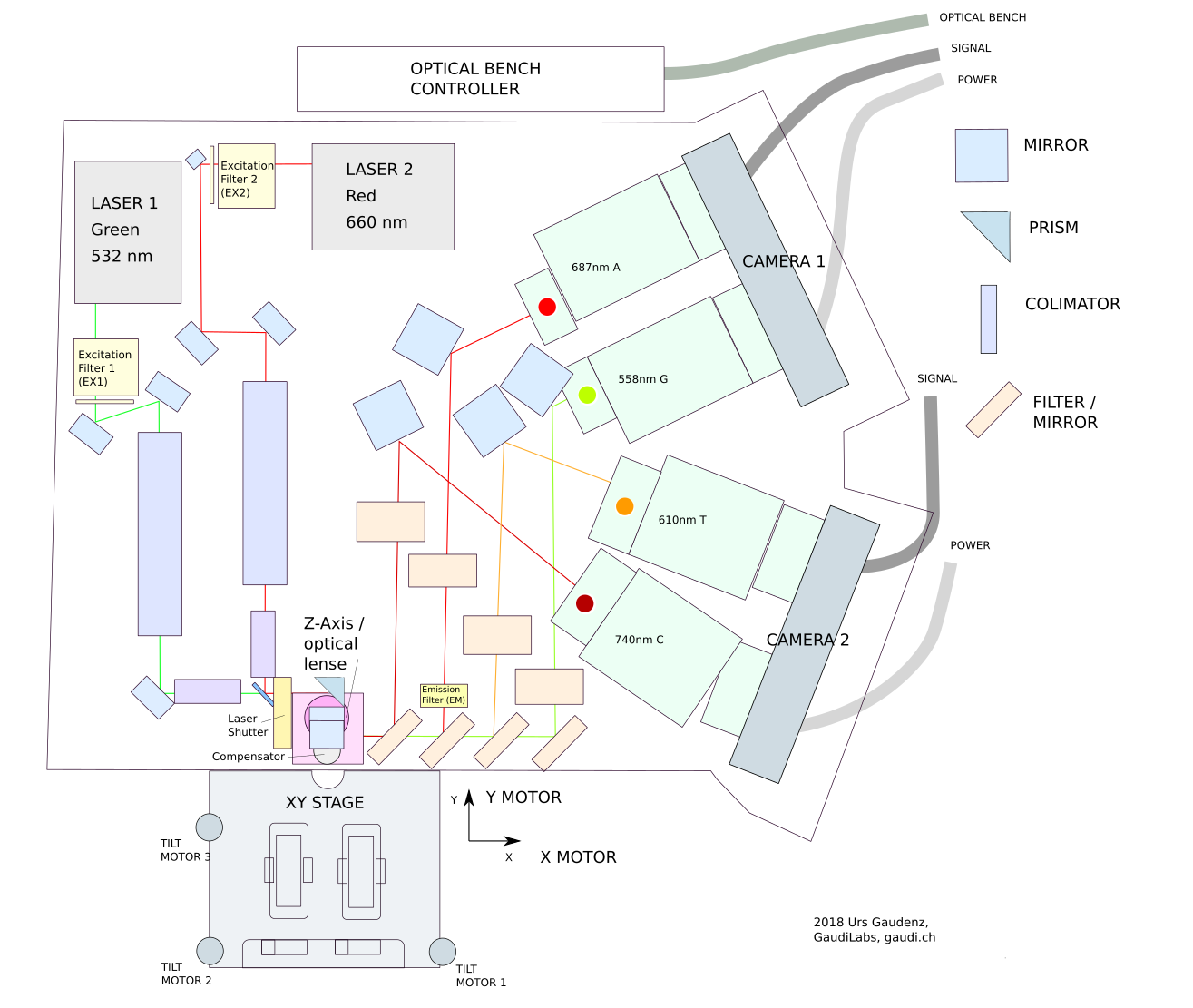
Oddly the most interesting thing here might be how available these systems are. It appears that they’re being replaced en masse and have become easily available in the range of thousands of dollars on the secondary market. At that price point they’re almost worth snapping up for the enclosure and parts! But we prefer Hackteria’s goal of enabling the Citizen Scientist to make use of these incredible machines for their intended purpose. Who knows what exciting projects we’ll find when hackers start sequencing their cats!
Thanks for the tip [kaspar]!
















Great article and phenomenal work. Thanks for writing this up!
DNA Sequencers have been the DIY laser hobbyists pick of choice for about ten years now (or more)… JSDU Argon Ion lasers are readily available and in my experience 30-50% are working out of the box… another 10-20% need some adjustments in the tube… some are just dead. I have been lucky and picked up a couple complete units with PSU, fan and head for around $100-$150 on ebay…. but have had to throw a couple in the bin as well.
Part of me wants to cringe at what they cost brand new. Anybody know?
Kaspar linked some blog posts to me that suggested around 500 or 700k. He also linked to a Craigslist post for $1,200….
If you only need to sequence one genome, MinION flow cells start at $500. They’re single use unfortunately.
How much of a genome? As in all 46 chromosomes? Or just one sequence of a few hundred base pairs?
The whole thing.
But gene sequencing Isn’t straight forward. You don’t just spit on the sensor and get your genes back. The sensor reads d/r-na fragments (whose lengths vary depending on the process) which then need to be sorted, aligned, & assembled. Fortunately there’s open source software you can run on your computer to assemble it. Something like E. coli might take a day on a mid range modern computer or a couple hours on a decent gaming rig. There’s a bunch of software built for gpus and server hardware so you might be able to sequence yourself in under a week if you’ve got the hardware.
It could be straight forward though. Read it like a single long tape by reading a single strand directly. Single molecule reads. Not sure we are “easily” able to do that quite yet but it’s technically possible to do so and starting to become commercially possible. Interestingly, our bodies do it all the time.
https://www.youtube.com/watch?v=wvorystm4A0
15kb (kilobase not kilobyte) is a long read but far from ‘reading it [the genome] like a single long tape’. For reference, E. coli is 4Gb long. Life unzips, snips, and matches pairs like a mariner splicing a rope or your torrent tracker sharing files (but without the fidelity); not like a film reel.
It’s getting there and physically possible but it’s not standard commercial practice right now is all. The process to unravel the entire genome is still done all the time when DNA is duplicated, which happens all the time. Nothing physically prevents some process from unraveling the entire sequence and reading the data of every single base pair.
*Mb
From the look of that diagram, it can see individual ATCG bases by looking at them!? Makes sense I suppose!
I wonder, with a few years’ hacking, if these could bring amateur competition into the “sequence your own DNA” market. The kits you send off a swab or whatever and they send you back meaningless statistics for you to overinterpret. They just look at a few particular sites on chromosomes AIUI. If you could do it cheaper, it might even be an income stream for hackerspaces. Certainly if it’s viable for companies to do it commercially, buying their equipment new and paying those hefty scientist wages!
Big problem might be getting certified though. Even though they’re not diagnostic or in any way used to treat illness, I suppose there’s probably some standards or other stopping companies just sending any old shit back to customers. Although it’d be pretty easy to get away with. For a while at least, until it gets cheap enough that whole families start doing it, and wondering why they don’t have any genes in common with their twin sisters.
Wonder if the machinery is accurate enough for holography? If you plopped it onto a very solid vibration-proof base. Bonkers, really, that there’s people reading this who grew up getting surplus WWII valve radio stuff from shops like that (bit older than me), and now there’s second-hand genetic sequencers going for not much more than scrap metal prices.
“They just look at a few particular sites on chromosomes AIUI. ”
It makes sense to do it that way, those particular sites are where most of the “ancestry” data lies.
I mean, who cares if your liver is a time bomb, if all you are interested in finding out where your ancestors came from.
B^)
You could probably scrape that data from coroners reports & obituaries with near the accuracy of some of these less scrupulous companies.
Can’t find any on Ebay for under a the price of a car. The cheap ones must have all been joinked after this article was published.
The E. Coli genome is about 5 Mb long, while the human genome is about 3.2 Gb. The Illumina instruments do not see single bps of DNA. Rather a large number of identical DNA molecules are in one spot, and the combined fluorescence signal of adding a labeled base to all the DNA molecules is detected.
There is a different DNA sequencing technology, nanopore sequencing, that reads the electrical signal of a single molecule of DNA passing through a small hole. Oxford Nanopore sells devices that use this method of DNA sequencing. The single molecule sequence reads from their machines can be 100’s of kb long.
“It appears that they’re being replaced en masse and have become easily available in the range of thousands of dollars on the secondary market. ”
The third world benefits. Trickle-down science.
Hi guys, I have a few of these machines for sale, if you’re interested in buying one let me know.. I’ll try to give you a good deal. Its the same model that is referenced in the article, the ILLUMINA HISEQ 2000.
Hey folks, thanks for everyone’s interest. The campaign to write the software for these machines has been live for over a month now and we just have 6 days left! Could really use your support.
https://wemakeit.com/projects/reseq-reuse-dna-sequencers?locale=en
Wish someone would tear down ADVA and change CART cancer therapies. CART cost $500K today.
https://vimeo.com/304435395
CART
https://www.junotherapeutics.com/the-science/our-platform/
Chimeric antigen receptor (CAR) T cell therapy is a relatively new and in many cases highly effective immunotherapy treatment for certain types of cancer. Obviously it comes at a cost, however: The manufacturers charge up to EUR 380,000 – 420.000 for the production of immune cells (generically modified ) for a single patient. This is why, in order to decrease the CAR-T cost, MIRCOD actively participates in R&D projects initiated by international consortium. Gene therapy technology (Fully automatic Point of Care drug manufacturing station) developed in consideration with international top cell and gene biotech companies and universities (Orgenesis, At-Vio, UC Davis etc..), guarantees to transform tomorrow’s medicine by treating the causes of disease rather than the manifestations. Based on initial results and tests, we estimate the commercial cost for CAR-T manipulations, on a new equipment to be around 60.000 to 80.000 USD per patient.