When exploring the realm of Machine Learning, it’s always nice to have some real and interesting data to work with. That’s where the bats come in – they’re fascinating animals that emit very particular ultrasonic calls that can be recorded and analysed with computer software to get a fairly good idea of what species they are. When analysed with an FFT spectogram, we can see the individual call shapes very clearly.
Creating an open source classifier for bats is also potentially useful for the world outside of Machine Learning as it could not only enable us to more easily monitor bats themselves, but also the knock on effects of modern farming methods on the natural environment. Bats feed on moths and other night flying insects which themselves have been decimated in numbers. Even in the depths of the countryside here in the UK these insects are a fraction of the population that they used to be 30 years ago, but nobody seems to have monitored this decline.
So getting back to our spectograms, it would be perfectly reasonable to throw these images at a convolutional neural network (CNN) and use an image feature-recognition strategy. But I wanted to explore the depths of the mysterious Random Forest.
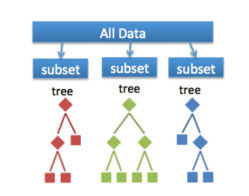 And what exactly is a Random Forest? As expected, the forest is comprised of trees, decision trees. It’s like playing a game of twenty questions. Sequential decisions are made, and each of these rule out some possible classifications. If each game of twenty questions is a tree, the Random Forest (RF) in the example below is composed of 4,000 trees and is like asking 4,000 people to play the game for you. We apply some kind of averaging formula to collate all the answers together and get a method for making the final decision that’s markedly better than if we’d done it alone.
And what exactly is a Random Forest? As expected, the forest is comprised of trees, decision trees. It’s like playing a game of twenty questions. Sequential decisions are made, and each of these rule out some possible classifications. If each game of twenty questions is a tree, the Random Forest (RF) in the example below is composed of 4,000 trees and is like asking 4,000 people to play the game for you. We apply some kind of averaging formula to collate all the answers together and get a method for making the final decision that’s markedly better than if we’d done it alone.
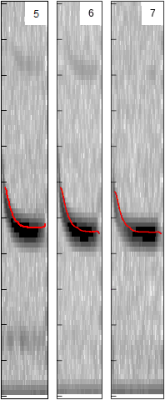 When you play twenty questions, you ask about the defining features of the thing in question. The obvious place to start is the point in the call where the call is the loudest (shown in black in image on left), which would give us a frequency. So in the case of the brown eared bat, the number would be something like 30 kHz. We don’t even have to call it anything else – the trees will just process blindly all the numbers in a column of data not needing to know what they mean in the real world. However, as sentient beings, we can actually see the data if we want to and even see what features of the spectogram were most important. Maybe the maximum amplitude frequency is also important? We don’t know yet – we’ll let the decision trees decide that for themselves. The red line in the image on the left shows the distinctive shape of the pipistrelle echo location call, filtered by the software to produce a nice thin line from which we can now derive values for things such as upward slope and steepness.
When you play twenty questions, you ask about the defining features of the thing in question. The obvious place to start is the point in the call where the call is the loudest (shown in black in image on left), which would give us a frequency. So in the case of the brown eared bat, the number would be something like 30 kHz. We don’t even have to call it anything else – the trees will just process blindly all the numbers in a column of data not needing to know what they mean in the real world. However, as sentient beings, we can actually see the data if we want to and even see what features of the spectogram were most important. Maybe the maximum amplitude frequency is also important? We don’t know yet – we’ll let the decision trees decide that for themselves. The red line in the image on the left shows the distinctive shape of the pipistrelle echo location call, filtered by the software to produce a nice thin line from which we can now derive values for things such as upward slope and steepness.
Extracting the relevant features like this is incredibly important. A data scientist, in conjunction with a bat call expert, would now look at the whole range of bat calls and make educated guesses as to which features are worth looking at – maybe the way the call slopes or if it starts with a ‘bang’ of amplitude or not? They might produce several hundred different columns for the data set and prune them down to about twenty, discarding all the ones that the trees themselves decide not to use very much.
Bring in the Bat Experts
After a few false starts, I finally settled on this system authored by Jean Marchal, Francois Fabianek and Christopher Scott. It’s basically a feature extraction package written in R that has been specifically targeted at bat and bird calls. There’s an easy to follow tutorial that downloads bird data and classifies one single bird and as someone of absolutely no previous experience with R, I was easily able to use it to cobble together a multi-class version that classified all the six species of bat that live near my home. I’m sure that there’s a more elegant way to implement in Random Forest, but I just classified each species separately and then combined the results. Please feel free to improve my method, but it does seem to work well.
The system is best run in Rstudio on Windows 10 and the whole thing installs seamlessly from start to finish. I also got it working in Ubuntu 18, but only from the command line using ‘$ Rscript Train_bats.R‘ . I never actually completed the official tutorial and found the data structure rather confusing to begin with. Thankfully, it turned out to be very simple. It seems that the system I created is a series of ‘binary classifiers’ that spits out a new confusion matrix for each species, which is trained against all the other species in the ‘data’ directory.
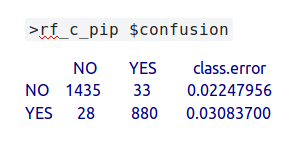
The species under investigation is the common pipistrelle – c_pip. During the automatic training and testing process, 1435 data points that were actually ‘not c_pip’ were correctly predicted as being ‘not c_pip’ … which is good! Furthermore, 880 labels which were actually c_pip were correctly predicted to be c_pip, which is also good. Unfortunately, 33 of the actual c_pip bats were wrongly predicted as not being that species and, even worse, 28 that were actually not c_pip were incorrectly predicted as being c_pip – damn! Thankfully, the percentages of incorrect predictions are fairly low – 2.2% and 3.1% respectively, in spite of only having 320 MB of bat data.
The results may be improved by working with more data, so if you’ve got any full spectrum .wav files lurking on your USB drives for the 17 species of UK bats, please send them to me! The recordings need to be in the order of 384 ks per sec (192 KHz) and not be distorted / clipped. Faint recordings are OK as they represent real life conditions. The other thing to mention is that the same species of bat in a different geographical location may well have different calls so it’s important to add one’s own local bat calls to the data for good results.
My own data is offered license free from Google Drive here. Use it any way you see fit! The species were identified with the help of experts in bat echo location on the Facebook group: Bat Call Sound Analysis Workshop. (Thanks guys!)
So we’ve got about 320 MB of training data, a working classifier in Rstudio and a recording of a bat from the previous evening using an UltraMic384K mic. It’s now simply a case of finding the directory Bioacoustics/unknown_bat_audio/, deleting any existing files and pasting in our unknown bat. Simple! Hit the ‘run’ button ……..
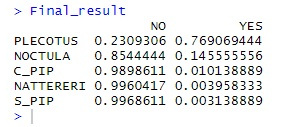 It’s very probably a plecotus auritus, the eminently cute brown long eared bat! Sometimes the classifier does struggle – particularly if the recordings are indistinct. Also, bats within the Myotis genus tend to have very similar calls to each other and actually overlap so are often impossible to classify without physically catching the creature and extracting DNA, or such like. This is not recommended by myself and if you really must do it, check if a license from the Feds is required and don’t get bitten – bats can carry some deadly diseases.
It’s very probably a plecotus auritus, the eminently cute brown long eared bat! Sometimes the classifier does struggle – particularly if the recordings are indistinct. Also, bats within the Myotis genus tend to have very similar calls to each other and actually overlap so are often impossible to classify without physically catching the creature and extracting DNA, or such like. This is not recommended by myself and if you really must do it, check if a license from the Feds is required and don’t get bitten – bats can carry some deadly diseases.
So what’s next? Maybe port the classifier to a Raspberry Pi and send the results out through a LoRa connection? Or if there’s a 2G cell phone signal, send it via HTTP? Or a hand held bat detector that speaks out the name of the bat. “Hey folks. There goes Barry the Brown Eared Bat, and he’s in the mood to party!”

















But if you were bitten and has been feeling some aversion to sunlight and garlic, then start negotiating the payments for your new castle. Don´t worry, you can mortgage it for a couple centuries….
The bat conservation trust (UK) have a library of bat calls – don’t know if thats the data set you already have? If not it might be worth getting in touch…
This a great project and is the kind of thing the natural world needs. Thanks and well done!
Despite being a major mammal group bats are very difficult to observe and are drastically underrepresented in nature studies. I suggest the following future step:
A possible direction is long term recording at various sites with automated identification and upload to iNaturalist via their API. Any observations that qualify as “research grade” are then passed on to the Global Biodiversity Information Facility (gbif.org) where information is shared amongst major institutions worldwide for research purposes. Both websites feature various tools for mapping and data visualization.
Thanks for the thumbs up! That’s a great idea re iNaturalist. One of the main problems is the cost of the equipment for full spectrum audio although there is an audio project based on teensy 3.6 which looks very promising. My own idea is to port it to raspberry pi 4 with a relatively cheap digital mic. eg SPH0641LU4H-1 .