Display technology has come a long way since the advent of the CRT in the late 1800s (yes, really!). Since then, we’ve enjoyed the Nixie tubes, flip dots, gas plasma, LCD, LED, ePaper, the list goes on. Now, there’s a new kid on the block — water.
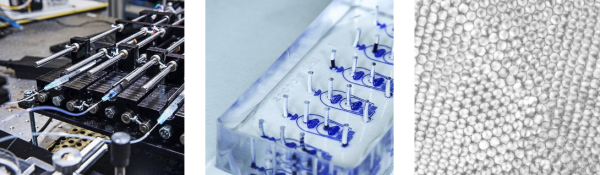
[Steve Mould] recently got his hands on an OpenDrop — an open-source digital microfluidics platform for biology research. It’s essentially a grid of electrodes coated in a dielectric. Water sits atop this insulating layer, and due to its polarized nature, droplets can be moved around the grid by voltages applied to the electrodes. The original intent was to automate experiments (see 8:19 in the video below for some wild examples), but [Steve] had far more important uses in mind.
When [Steve]’s €1,000 device shipped from Switzerland, it was destined for greatness. It was turned into a game console for classics such as Pac-Man, Frogger, and of course, Snake. With help from the OpenDrop’s inventor (and Copilot), he built paired-down versions of the games that could run on the 8×14 “pixel” grid. Pac-Man in particular proved difficult, because due to the conservation of mass, whenever Pac-Man ate a ghost, he grew and eventually became unwieldy. Fortunately, Snake is one of the few videogames that actually respects the laws of classical mechanics, as the snake grows by one unit each time it consumes food.
[Steve] has also issued a challenge — if you code up another game, he’ll run it on his OpenDrop. He’s even offering a prize for the first working Tetris implementation, so be sure to check out his source code linked in the video description as a starting point. We’ve seen Tetris on oscilloscopes and 3D LED matrices before, so it’s about time we get a watery implementation.