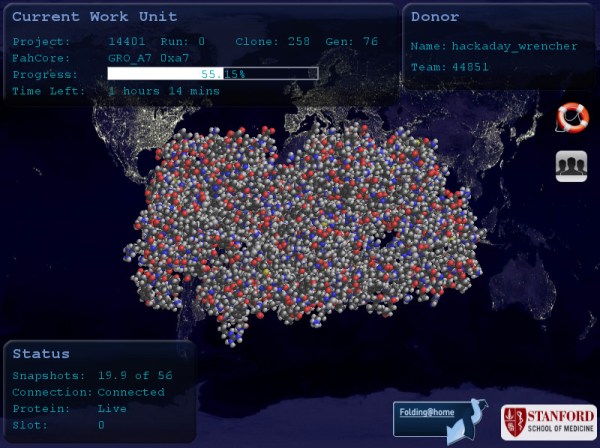
A great big Thank You to everyone who answered the call to participate in Folding@Home, helping to understand proteins interactions of SARS-CoV-2 virus that causes COVID-19. Some members of the FAH research team hosted an AMA (Ask Me Anything) session on Reddit to provide us with behind-the-scenes details. Unsurprisingly, the top two topics are “Why isn’t my computer doing anything?” and “What does this actually accomplish?”
The first is easier to answer. Thanks to people spreading the word — like the amazing growth of Team Hackaday — there has been a huge infusion of new participants. We could see this happening on the leader boards, but in this AMA we have numbers direct from the source. Before this month there were roughly thirty thousand regular contributors. Since then, several hundred thousands more started pitching in. This has overwhelmed their server infrastructure and resulted in what’s been termed a friendly-fire DDoS attack.
The most succinct information was posted by a folding support forum moderator.
Here’s a summary of current Folding@Home situation :
* We know about the work unit shortage
* It’s happening because of an approximately 20x increase in demand
* We are working on it and hope to have a solution very soon.
* Keep your machines running, they will eventually fold on their own.
* Every time we double our server resources, the number of Donors trying to help goes up by a factor of 4, outstripping whatever we do.
Why don’t they just buy more servers?
The answer can be found on Folding@Home donation FAQ. Most of their research grants have restrictions on how that funding is spent. These restrictions typically exclude capital equipment and infrastructure spending, meaning researchers can’t “just” buy more servers. Fortunately they are optimistic this recent fame has also attracted attention from enough donors with the right resources to help. As of this writing, their backend infrastructure has grown though not yet caught up to the flood. They’re still working on it, hang tight!
Computing hardware aside, there are human limitations on both input and output sides of this distributed supercomputer. Folding@Home need field experts to put together work units to be sent out to our computers, and such expertise is also required to review and interpret our submitted results. The good news is that our contribution has sped up their iteration cycle tremendously. Results that used to take weeks or months now return in days, informing where the next set of work units should investigate.
Continue reading “Behind The Scenes Of Folding@Home: How Do You Fight A Virus With Distributed Computing?” →