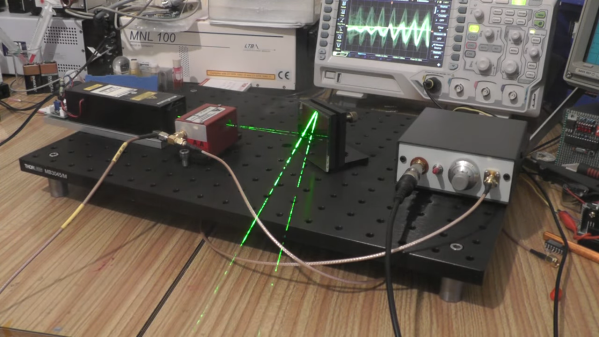
We all know that light and sound are wave phenomena, but of very different kinds. Light is electromechanical in nature, while sound is mechanical. Light can travel through a vacuum, while sound needs some sort of medium to transmit it. So it would seem that it might be difficult to use sound to modify light, but with the right equipment, it’s actually pretty easy.
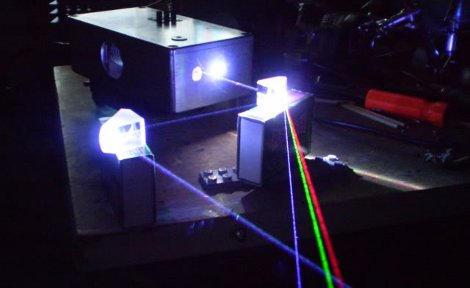
Easy, perhaps, if you’re used to slinging lasers around and terms like “acousto-optic tunable filter” fall trippingly from your tongue, as is the case for [Les Wright]. An AOTF is a device that takes a radio frequency input and applies it to a piezoelectric transducer that’s bonded to a crystal of tellurium oxide. The RF signal excites the transducer, which vibrates the TeO2 crystal and sets up a standing wave within it. The alternating bands of compressed and expanded material within the crystal act like a diffraction grating. Change the excitation frequency, and the filter’s frequency changes too.
To explore the way sound can bend light, [Les] picked up a commercial AOTF from the surplus market. Sadly, it didn’t come with the RF driver, but no matter — a few quick eBay purchases put the needed RF generator and power amplifier on his bench. The modules went into an enclosure to make the driver more of an instrument and less of a one-off, with a nice multi-turn pot and vernier knob for precise filter adjustment. It’s really kind of cool to watch the output beam change colors at the twist of a knob, and cooler still to realize how it all works.
We’ve been seeing a lot of [Les]’ optics projects lately, from homemade TEA lasers to blasting the Bayer filter off a digital camera, each as impressive as the last! Continue reading “Acousto-Optic Filter Uses Sound To Bend Light”