On Wednesday morning we asked the Hackaday community to donate their extra computer cycles for Coronavirus research. On Thursday morning the number of people contributing to Team Hackaday had doubled, and on Friday it had doubled again. Thank you for putting those computers to work in pursuit of drug therapies for COVID-19.
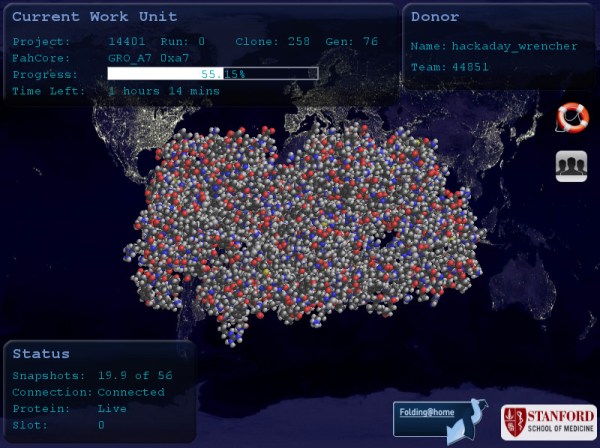
I’m writing today for two reasons, we want to keep up this trend, and also answer some of the most common questions out there. Folding@Home (FAH) is an initiative that simulates proteins associated with several diseases, searching for indicators that will help medical researchers identify treatments. These are complex problems and your efforts right now are incredibly important to finding treatments faster. FAH loads the research pipeline, generating a data set that researchers can then follow in every step of the process, from identifying which chemical compounds may be effective and how to deliver them, to testing they hypothesis and moving toward human trials.
Continue reading “Coronavirus And Folding@Home; More On How Your Computer Helps Medical Research”





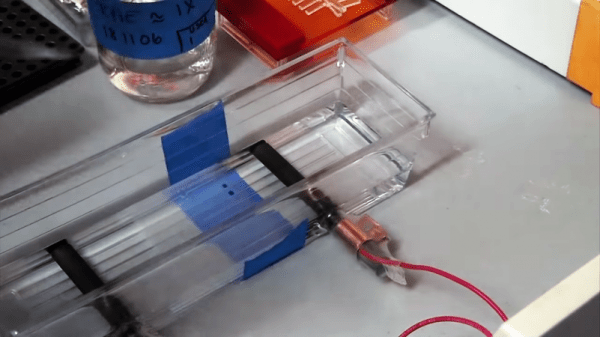
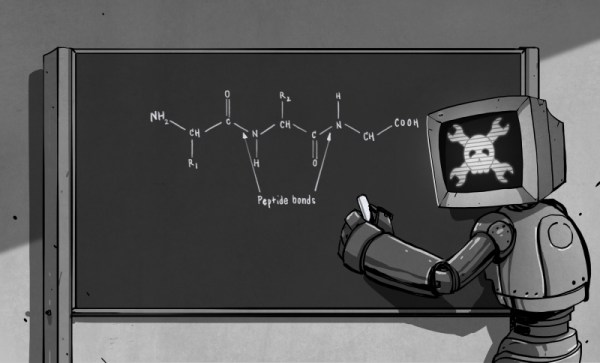

 For “Tears”, his sculpture of the enzyme lysozyme shown in the banner image, [Mike] started with crystallographic data that pinpoints every peptide residue in the protein. A model is created for the 3D printer, with careful attention paid to how the finished print can be split apart to allow casting. Clear PLA filament is used for the positive because it burns out of the mold better than colored plastic. The prints are solvent smoothed, sprues and air vents added, and the positive is coated with a plaster mix appropriate for the sculpture medium before the plastic is melted out and the mold is ready for casting.
For “Tears”, his sculpture of the enzyme lysozyme shown in the banner image, [Mike] started with crystallographic data that pinpoints every peptide residue in the protein. A model is created for the 3D printer, with careful attention paid to how the finished print can be split apart to allow casting. Clear PLA filament is used for the positive because it burns out of the mold better than colored plastic. The prints are solvent smoothed, sprues and air vents added, and the positive is coated with a plaster mix appropriate for the sculpture medium before the plastic is melted out and the mold is ready for casting.