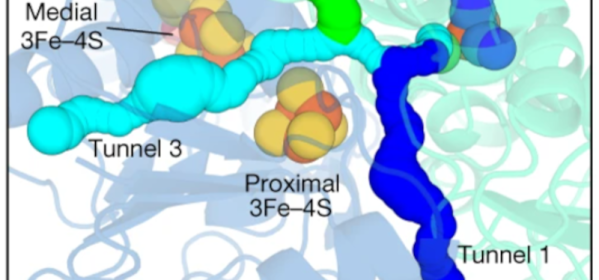
We’ve all been there — you forgot your lunch, but there are AC outlets galore. Wouldn’t it be so much simpler if you could just plug in like your phone? Don’t try it yet, but biologists have taken us one step further to being able to fuel ourselves on those sweet, sweet electrons.
Using an “electrobiological module” of 3-4 enzymes, the amusingly named AAA (acid/aldehyde ATP) cycle regenerates ATP in biological systems directly from electricity. The process takes place at -0.6 V vs a standard hydrogen electrode (SHE), and is compatible with biological transcription/translation processes like “RNA and protein synthesis from DNA.”
The process isn’t dependent on any membranes to foul or more complicated sets of enzymes making it ideal for in vitro synthetic biology since you don’t have to worry about keeping as many components in an ideal environment. We’re particularly interested in how this might apply to DNA computing which we keep being promised will someday be the best thing since the transistor.
Maybe in the future we’ll all jack in instead of eating our daily food pill? If this all seems like something you’ve heard of before, but in reverse, maybe you’re thinking of microbial fuel cells.