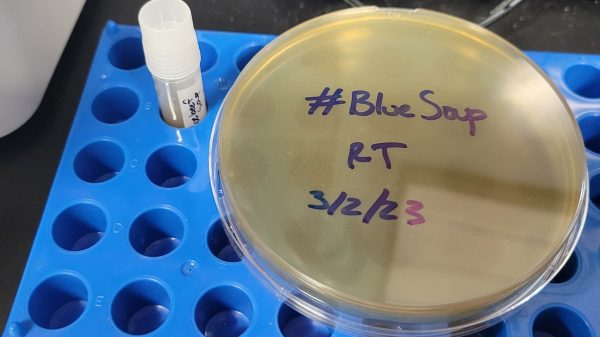
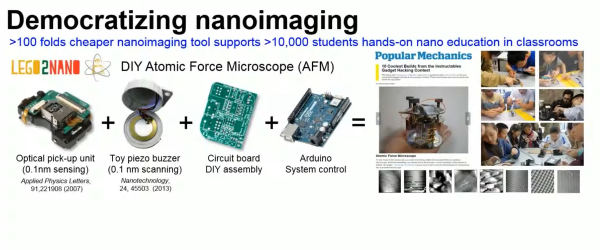
Beef soup! You’d normally expect it to be somewhere from reddish-brown to grey, depending on how well it was cooked and prepared. However, strangely, an assistant professor found the beef soup in their fridge had mysteriously turned blue. That spawned an investigation into the cause which is still ongoing.
[Dr. Elinne Becket] has earned her stripes in microbiology, but the blue soup astounded her. Despite her years of experience, she was unable to guess at the process or a source of contamination that could turn the soup blue. Indeed, very few natural foods are blue at all. Even blueberries themselves are more of a purple color. The case sparked enough interest that [Elinne] went back to the trash to collect photos and sample for research at the request of others.
Thus far, metagenomic DNA analysis is ongoing and samples of the soup have been cultivated in petri dishes. Early analysis shows that some of the microbes form iridescent colonies, Another researcher is trying to determine if the bugs from the soup can make blue color appear on soft cheese. There’s some suspicion that a bacteria known as pseudomonas aeruginosa could be the cause of the blue color, but that presents its own problems. P. aeruginosa is classified as a Biosafety Level 2 pathogen which would require some researchers to abandon work on the project for safety reasons.
The jury’s still out on this microbiological mystery. If you’ve got some ideas on what could be going on, let us know in the comments!